Search Count: 21
All
Selected
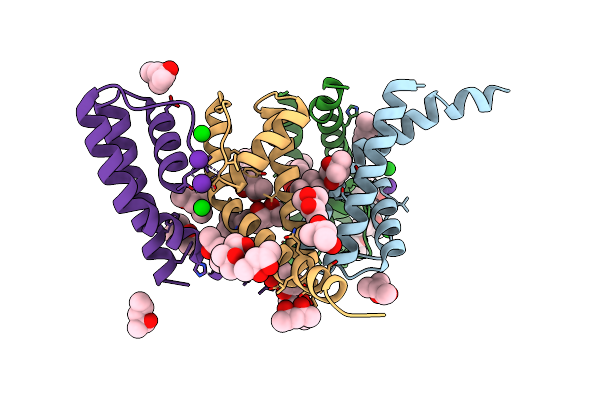 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN Ligands: MPD, RB, BA |
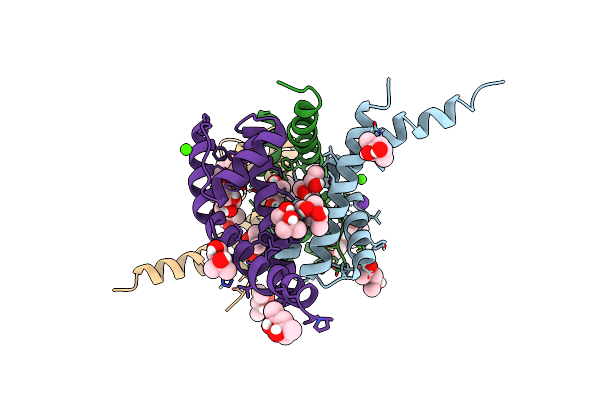 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN Ligands: RB, CA, MPD, CL |
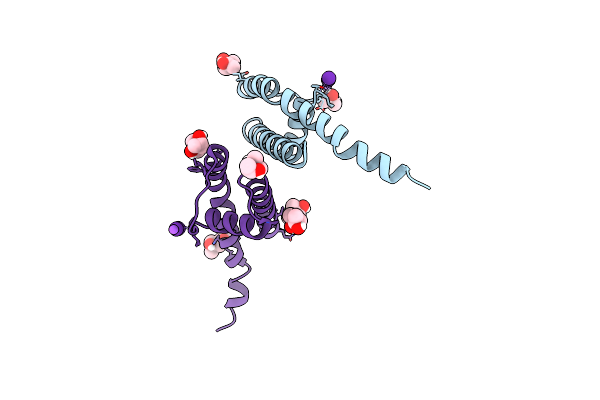 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-02-01 Classification: TRANSPORT PROTEIN Ligands: RB, MPD, NA |
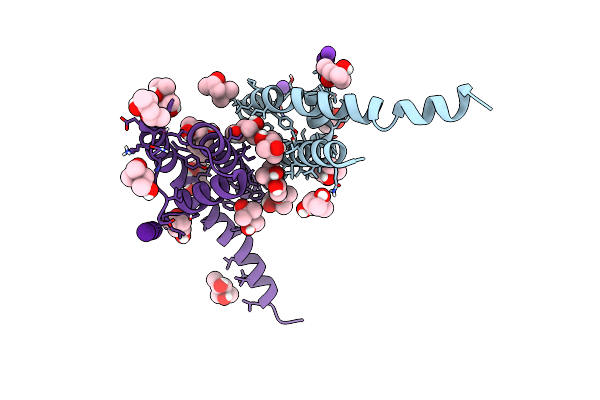 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-02-01 Classification: TRANSPORT PROTEIN Ligands: CS, MPD, K |
 |
Organism: Bacillus cereus (strain atcc 14579 / dsm 31 / jcm 2152 / nbrc 15305 / ncimb 9373 / nrrl b-3711)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-08-10 Classification: TRANSPORT PROTEIN Ligands: RB, MPD, K |
 |
Organism: Bacillus cereus (strain atcc 14579 / dsm 31 / jcm 2152 / nbrc 15305 / ncimb 9373 / nrrl b-3711)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-06-22 Classification: TRANSPORT PROTEIN Ligands: MPD, K, ACT, CL, MRD, NA |
 |
Organism: Bacillus cereus (strain atcc 14579 / dsm 31 / jcm 2152 / nbrc 15305 / ncimb 9373 / nrrl b-3711)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-06-22 Classification: TRANSPORT PROTEIN Ligands: MPD, NA, K, CL |
 |
Organism: Bacillus cereus (strain atcc 14579 / dsm 31 / jcm 2152 / nbrc 15305 / ncimb 9373 / nrrl b-3711)
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-06-08 Classification: TRANSPORT PROTEIN Ligands: K, MPD, MRD, NA, CL |
 |
Organism: Bacillus cereus (strain atcc 14579 / dsm 31 / jcm 2152 / nbrc 15305 / ncimb 9373 / nrrl b-3711)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-06-08 Classification: TRANSPORT PROTEIN Ligands: K, MPD, MRD, ACT, CL |
 |
Organism: Bacillus cereus (strain atcc 14579 / dsm 31 / jcm 2152 / nbrc 15305 / ncimb 9373 / nrrl b-3711)
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2022-06-08 Classification: TRANSPORT PROTEIN Ligands: K, MPD, CL, MRD |
 |
Crystal Structure Of The Glua2 Lbd (L483Y-N754S-L758V) In Complex With Glutamate
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2017-09-13 Classification: TRANSPORT PROTEIN Ligands: 1PE, PG0, SO4, NA, GLU |
 |
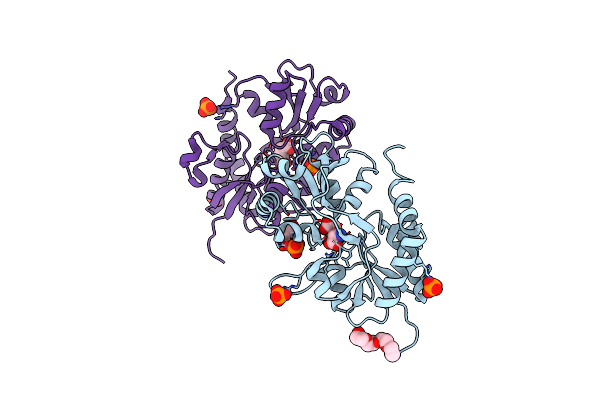
Crystal Structure Of A Tetramer Of Glua2 Tr Mutant Ligand Binding Domains Bound With Glutamate At 1.26 Angstrom Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2016-01-13 Classification: TRANSPORT PROTEIN Ligands: GLU, PO4, PEG |
 |
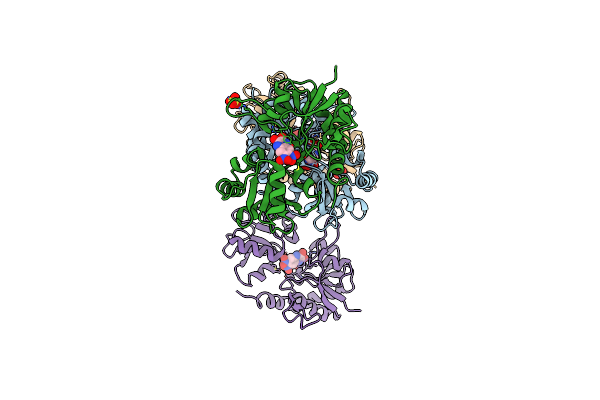
Crystal Structure Of A Tetramer Of Glua2 Ligand Binding Domains Bound With Glutamate At 1.45 Angstrom Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-01-13 Classification: TRANSPORT PROTEIN Ligands: GLU, PO4, PG4, PEG |
 |
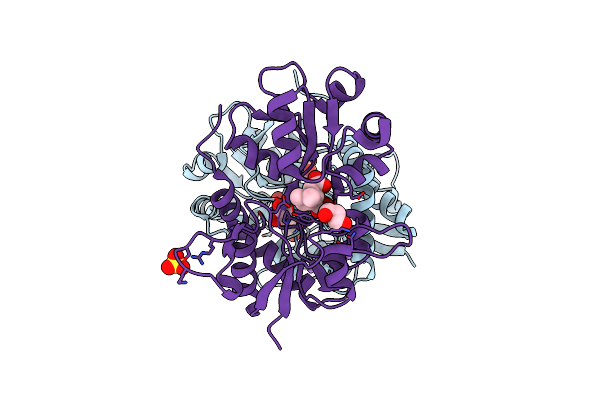
Glua2-L483Y-A665C Ligand-Binding Domain In Complex With The Antagonist Dnqx
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-08-14 Classification: TRANSPORT PROTEIN Ligands: DNQ, SO4 |
 |
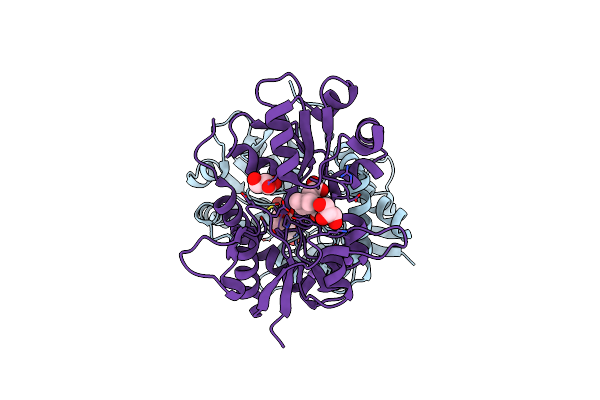
Crystal Structure Of Glur5 Ligand-Binding Core In Complex With Lithium At 1.49 Angstrom Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2008-06-17 Classification: MEMBRANE PROTEIN Ligands: LI, CL, KAI, GOL, SO4 |
 |
Crystal Structure Of Glur5 Ligand-Binding Core In Complex With Sodium At 1.72 Angstrom Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2008-06-17 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of Glur5 Ligand-Binding Core In Complex With Potassium At 1.78 Angstrom Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2008-06-17 Classification: MEMBRANE PROTEIN Ligands: K, CL, KAI, GOL |
 |
Crystal Structure Of Glur5 Ligand-Binding Core In Complex With Rubidium At 1.82 Angstrom Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2008-06-17 Classification: MEMBRANE PROTEIN Ligands: RB, CL, KAI, GOL |
 |
Crystal Structure Of Glur5 Ligand-Binding Core In Complex With Cesium At 1.97 Angstrom Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2008-06-17 Classification: MEMBRANE PROTEIN Ligands: CS, CL, KAI, GOL |
 |
Crystal Structure Of Glur5 Ligand-Binding Core In Complex With Ammonium Ions At 1.68 Angstrom Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2008-06-17 Classification: MEMBRANE PROTEIN Ligands: NH4, CL, KAI, GOL |

