Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
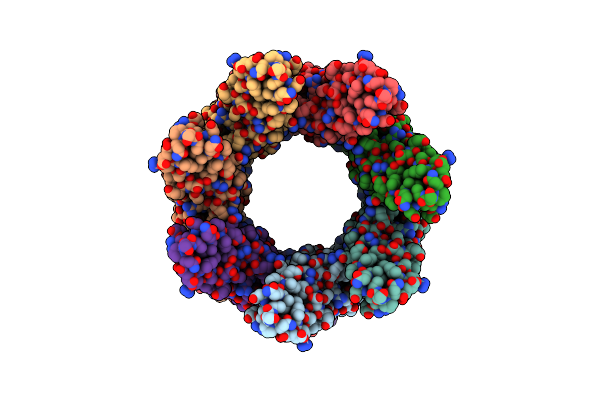 |
Cryoem Structure Of The Ring-Shaped Virulence Factor Espb From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: ELECTRON MICROSCOPY Release Date: 2020-06-17 Classification: TRANSPORT PROTEIN |
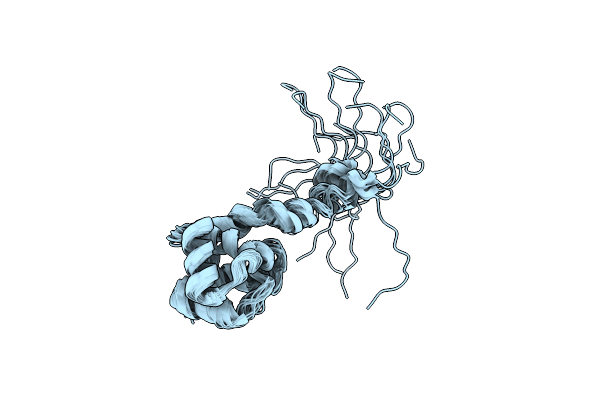 |
Structural And Dna Binding Properties Of Mycobacterial Integration Host Factor Mihf
Organism: Mycobacterium tuberculosis
Method: SOLUTION NMR Release Date: 2019-12-25 Classification: DNA BINDING PROTEIN |
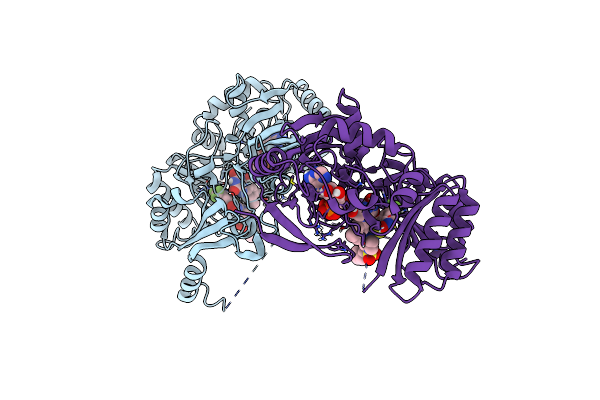 |
Crystal Structure Of M. Tuberculosis Dpre1 In Complex With Spbtz169 (Sulfonylpbtz)
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2018-08-01 Classification: OXIDOREDUCTASE Ligands: FAD, EQ8 |
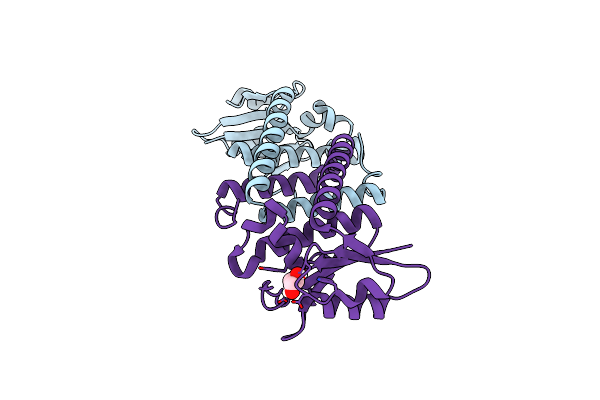 |
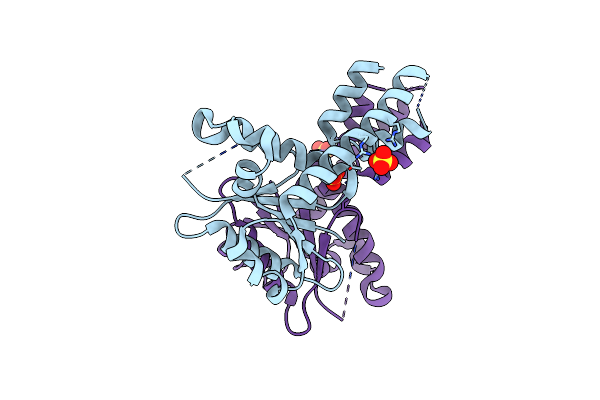
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-04-19 Classification: TRANSLATION Ligands: GOL |
 |
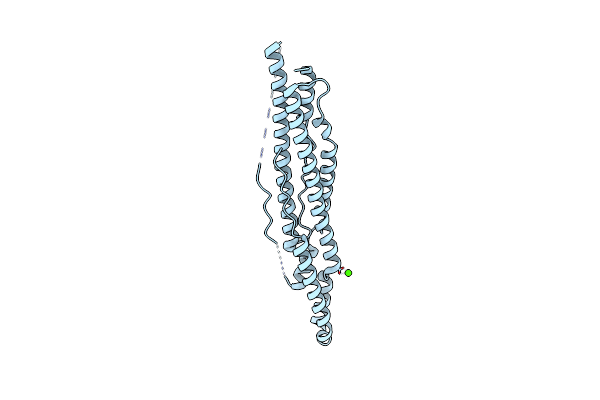
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:3.17 Å Release Date: 2017-04-19 Classification: TRANSLATION Ligands: SO4 |
 |
Structure Of Pe-Ppe Domains Of Esx-1 Secreted Protein Espb, C2221 In Presence Of Ca
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2015-02-18 Classification: PROTEIN TRANSPORT Ligands: CA |
 |
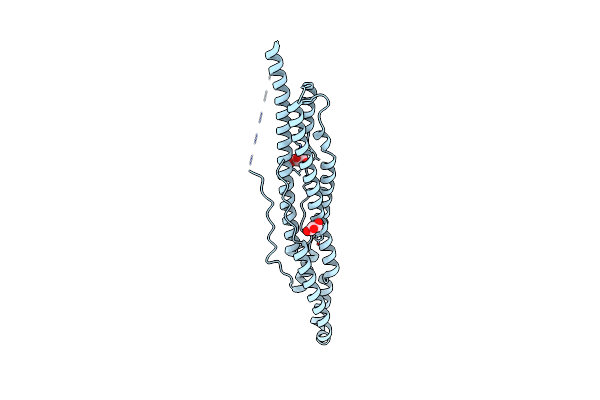
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2015-02-18 Classification: PROTEIN TRANSPORT Ligands: CL, NA |
 |
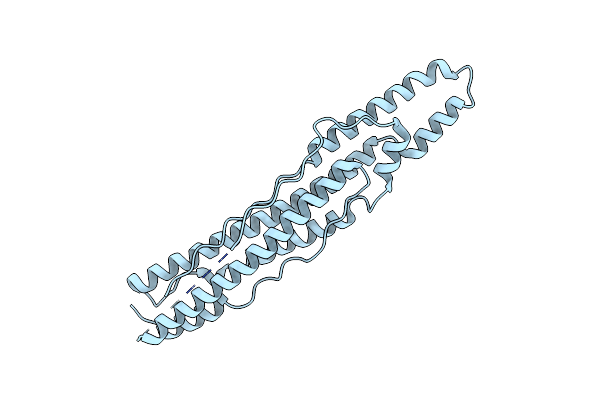
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-02-18 Classification: PROTEIN TRANSPORT Ligands: CL, GOL |
 |
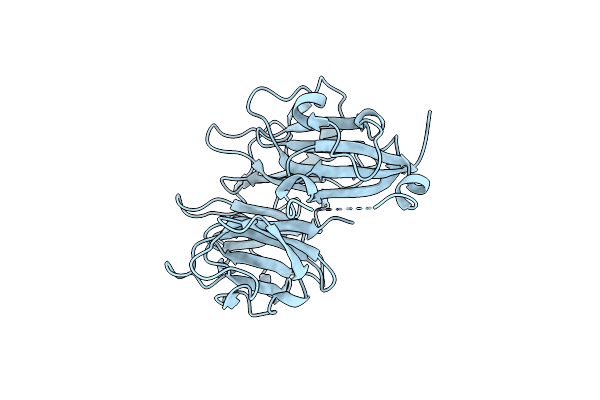
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2015-02-18 Classification: PROTEIN TRANSPORT |
 |
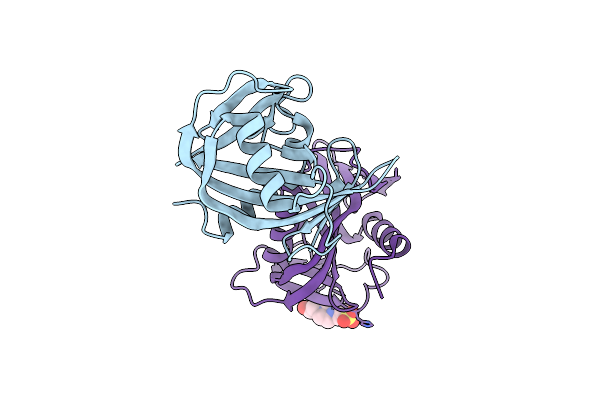
Mycobacterium Tuberculosis Gyrase Type Iia Topoisomerase C-Terminal Domain At 1.4 A Resolution
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-08-07 Classification: ISOMERASE |
 |
Crystal Structure Of The Bacillus Subtilis Pyrophosphohydrolase Bsrpph (E68A Mutant)
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-05-08 Classification: HYDROLASE Ligands: EPE |
 |
Crystal Structure Of The Bacillus Subtilis Pyrophosphohydrolase Bsrpph Bound To A Non-Hydrolysable Triphosphorylated Dinucleotide Rna (Pcp-Pgpg) - First Guanosine Residue In Guanosine Binding Pocket
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-05-08 Classification: HYDROLASE/RNA Ligands: EPE |
 |
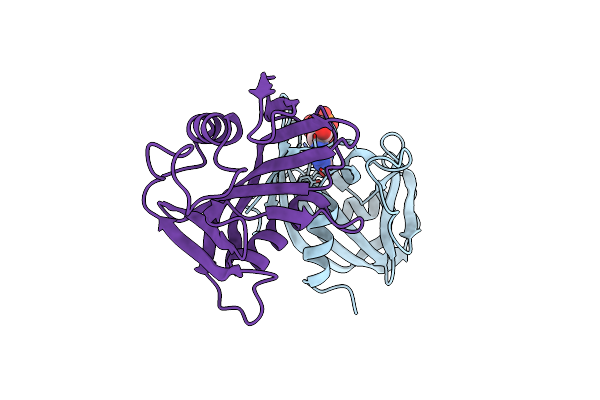
Crystal Structure Of The Bacillus Subtilis Pyrophosphohydrolase Bsrpph Bound To A Non-Hydrolysable Triphosphorylated Dinucleotide Rna (Pcp-Pgpg) - Second Guanosine Residue In Guanosine Binding Pocket
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-05-08 Classification: HYDROLASE/RNA Ligands: MG, EPE |
 |
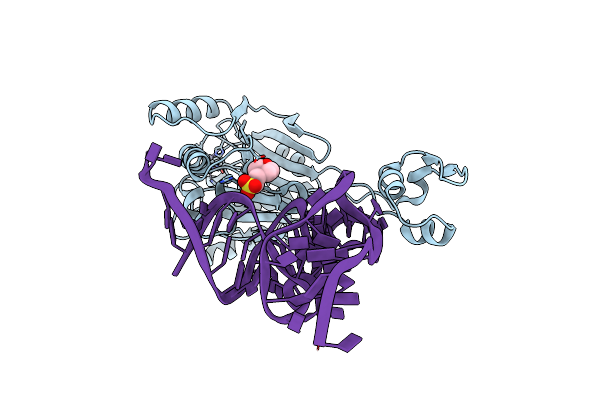
Crystal Structure Of The Bacillus Subtilis Pyrophosphohydrolase Bsrpph (E68A Mutant) Bound To Gtp
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2013-05-01 Classification: HYDROLASE Ligands: GTP |
 |
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-10-31 Classification: HYDROLASE/RNA Ligands: ZN, MES |
 |
Molecular Basis For The Recognition And Cleavage Of Rna (Uuccgu) By The Bifunctional 5'-3' Exo/Endoribonuclease Rnase J
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2011-10-19 Classification: HYDROLASE/RNA Ligands: ZN |
 |
Molecular Basis For The Recognition And Cleavage Of Rna (Cugg) By The Bifunctional 5'-3' Exo/Endoribonuclease Rnase J
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-10-19 Classification: HYDROLASE/RNA Ligands: ZN, GOL |
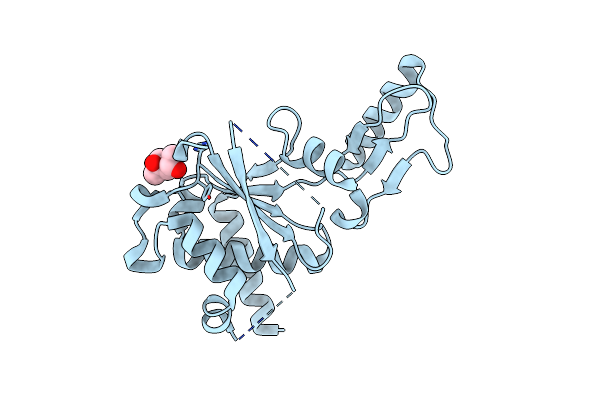 |
Crystal Structure Of The Second Part Of The Mycobacterium Tuberculosis Dna Gyrase Reaction Core: The Toprim Domain At 1.95 A Resolution
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-09-29 Classification: ISOMERASE Ligands: MPD |
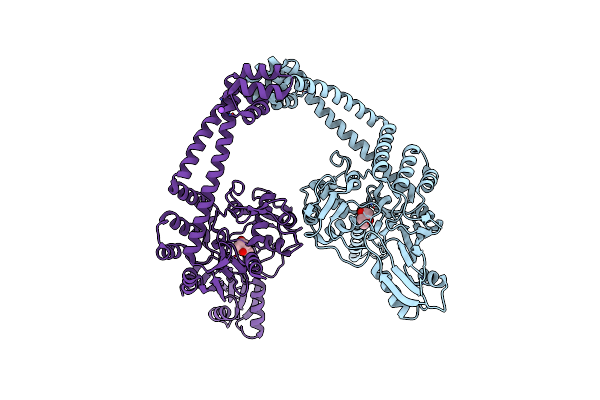 |
Crystal Structure Of The First Part Of The Mycobacterium Tuberculosis Dna Gyrase Reaction Core: The Breakage And Reunion Domain At 2.7 A Resolution
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-07-28 Classification: ISOMERASE Ligands: MPD, NA |
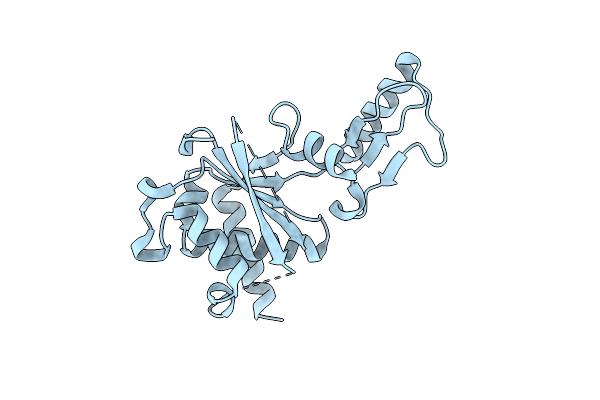 |
Crystal Structure Of The Second Part Of The Mycobacterium Tuberculosis Dna Gyrase Reaction Core: The Toprim Domain At 2.1 A Resolution
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-07-28 Classification: ISOMERASE |