Search Count: 16
 |
Organism: Myxococcus xanthus
Method: SOLUTION NMR Release Date: 2023-04-19 Classification: UNKNOWN FUNCTION |
 |
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2022-05-11 Classification: PROTEIN BINDING Ligands: PGE, EDO, PEG |
 |
Unbound State Of A De Novo Designed Protein Binder To The Human Interleukin-7 Receptor
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-05-11 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of The De Novo Designed Binding Protein H3Mb In Complex With The 1968 Influenza A Virus Hemagglutinin
Organism: Influenza a virus (strain a/hong kong/1/1968 h3n2), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-05-04 Classification: VIRAL PROTEIN/DE NOVO PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-04-20 Classification: DE NOVO PROTEIN Ligands: NAG, EDO, SO4 |
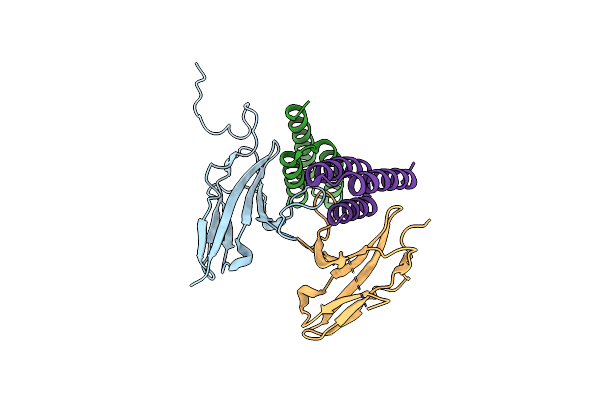 |
Crystal Structure Of Fgfr4 Domain 3 In Complex With A De Novo-Designed Mini-Binder
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2022-04-06 Classification: SIGNALING PROTEIN |
 |
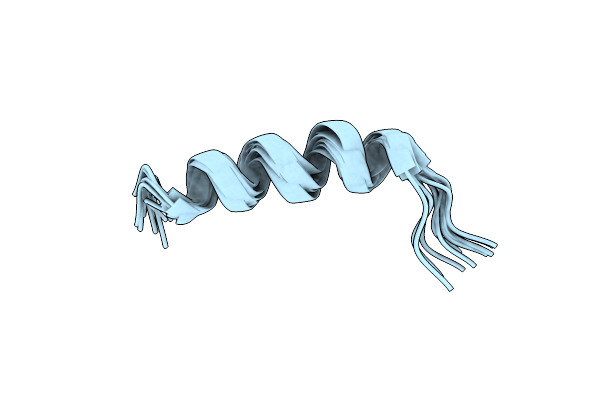
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2022-04-06 Classification: DE NOVO PROTEIN |
 |
Suboptimization Of A Glycine Rich Peptide Allows The Combinatorial Space Exploration For Designing Novel Antimicrobial Peptides
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2018-03-07 Classification: ANTIMICROBIAL PROTEIN |
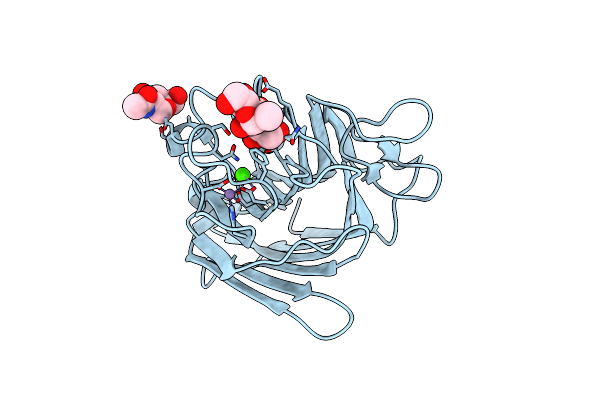 |
Crystal Structure Of Native Lectin From Platypodium Elegans Seeds (Pela) Complexed With Man1-3Man-Ome.
Organism: Platypodium elegans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-10-04 Classification: SUGAR BINDING PROTEIN Ligands: NAG, MN, CA |
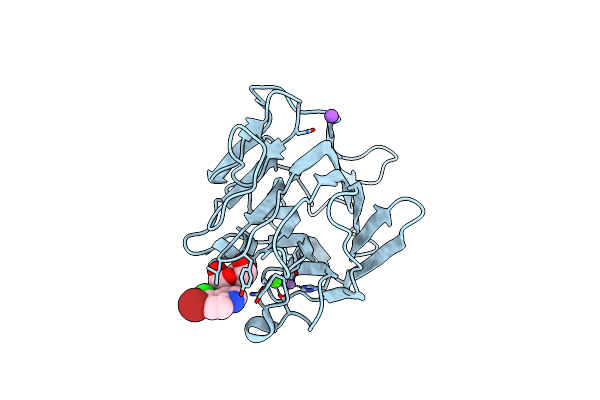 |
Organism: Cymbosema roseum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-10-01 Classification: Carbohydrate-binding protein Ligands: MN, CA, XMM, NA |
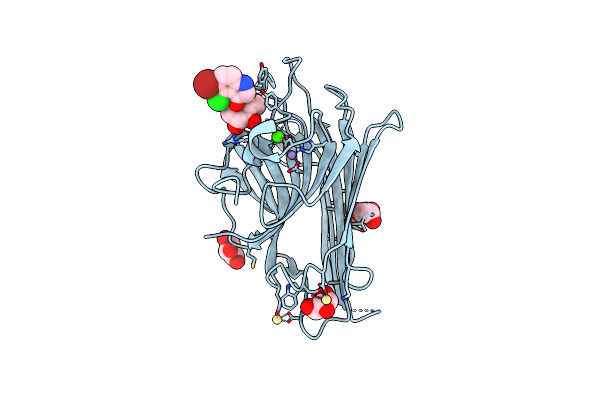 |
Crystal Structure Of Canavalia Grandiflora Seed Lectin Complexed With X-Man.
Organism: Canavalia grandiflora
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-05-21 Classification: sugar binding protein, plant protein Ligands: CA, MN, XMM, SO4, CD, GOL |
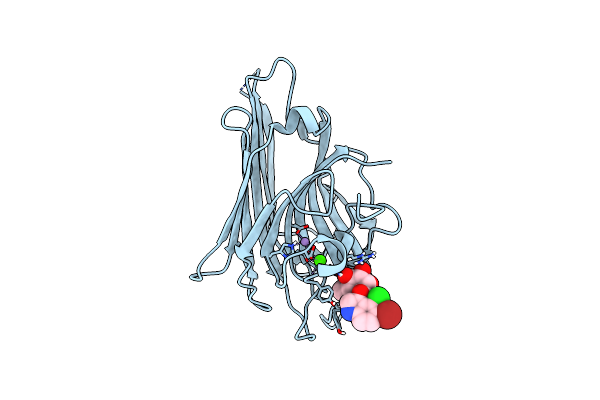 |
Three-Dimensional Structure Of Lectin From Dioclea Violacea And Comparative Vasorelaxant Effects With Dioclea Rostrata
Organism: Dioclea violacea
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2012-04-04 Classification: SUGAR BINDING PROTEIN Ligands: MN, CA, XMM |
 |
Organism: Dioclea virgata
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-02-08 Classification: SUGAR BINDING PROTEIN Ligands: MN, CA, XMM |
 |
Organism: Dioclea virgata
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2011-11-09 Classification: carbohydrate binding protein Ligands: MN, CA |
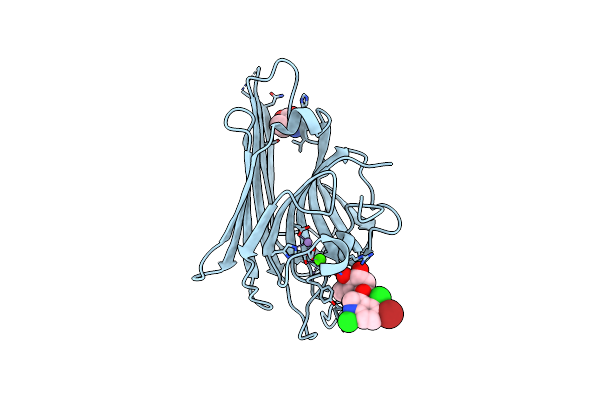 |
Crystal Structure Of A Pro-Inflammatory Lectin From The Seeds Of Dioclea Wilsonii Standl
Organism: Dioclea wilsonii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-10-05 Classification: Carbohydrate binding protein Ligands: MN, CA, XMM, CL, A3B |
 |
Crystal Structure Of An Antiflamatory Legume Lectin From Cymbosema Roseum Seeds
Organism: Cymbosema roseum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-04-07 Classification: SUGAR BINDING PROTEIN Ligands: MN, CA, ABA |

