Search Count: 17
 |
Crystal Structure Of Mgat5 Bump-And-Hole Mutant In Complex With Udp And M592
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-10-02 Classification: TRANSFERASE Ligands: UDP, NAG, MAN, SO4 |
 |
Organism: Aggregatibacter aphrophilus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-09-06 Classification: TRANSFERASE Ligands: EDO, GDU |
 |
Organism: Aggregatibacter aphrophilus
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2023-09-06 Classification: TRANSFERASE Ligands: U2F, UDP |
 |
Organism: Aggregatibacter aphrophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-09-06 Classification: TRANSFERASE Ligands: UDP |
 |
Crystal Structure Of Txgh116 D593A Acid/Base Mutant From Thermoanaerobacterium Xylanolyticum
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-03 Classification: HYDROLASE Ligands: GOL, CA |
 |
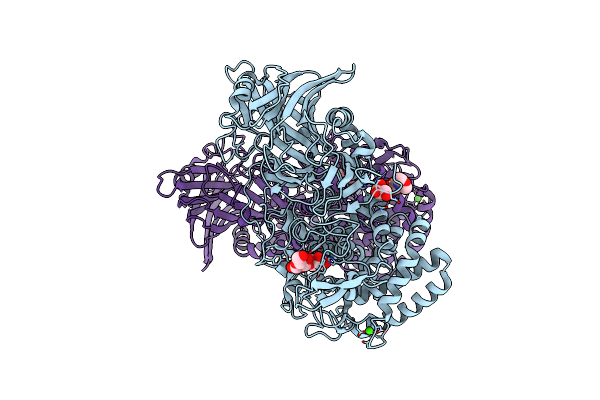
Crystal Structure Of Txgh116 D593A Acid/Base Mutant From Thermoanaerobacterium Xylanolyticum With Cellobiose
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-05-03 Classification: HYDROLASE Ligands: GOL, CA, MES |
 |
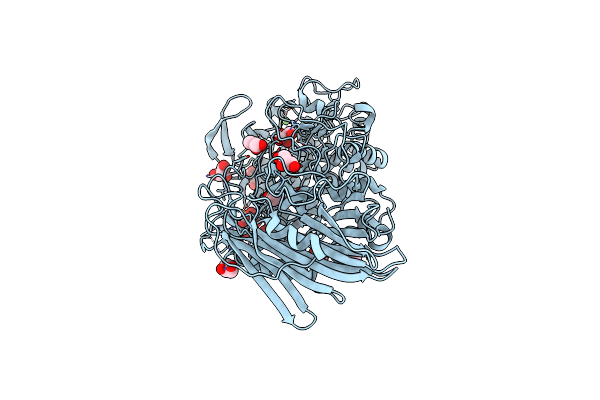
Crystal Structure Of Txgh116 D593A Acid/Base Mutant From Thermoanaerobacterium Xylanolyticum With Laminaribiose
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-03 Classification: HYDROLASE Ligands: CA, GOL, MES |
 |
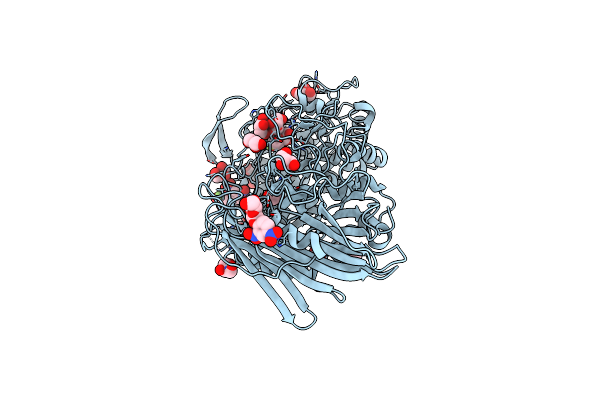
Crystal Structure Of Txgh116 D593N Acid/Base Mutant From Thermoanaerobacterium Xylanolyticum
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-05-03 Classification: HYDROLASE Ligands: GOL, CA, EDO |
 |
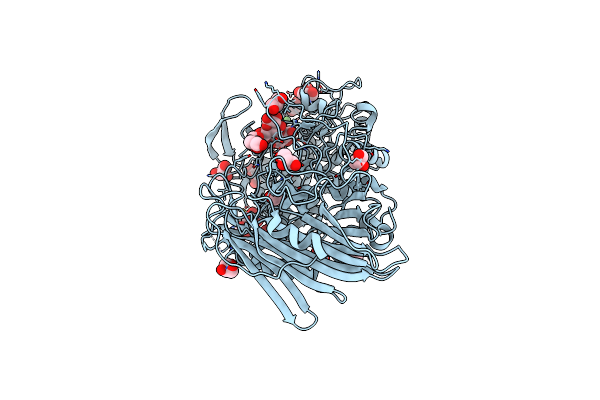
Crystal Structure Of Txgh116 D593N Acid/Base Mutant From Thermoanaerobacterium Xylanolyticum With 2-Deoxy-2-Fluoroglucoside
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-05-03 Classification: HYDROLASE Ligands: G2F, GOL, CA, EDO, NFG |
 |
Crystal Structure Of Txgh116 D593N Acid/Base Mutant From Thermoanaerobacterium Xylanolyticum With Cellobiose
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-05-03 Classification: HYDROLASE Ligands: GOL, CA, EDO |
 |
Crystal Structure Of Txgh116 D593N Acid/Base Mutant From Thermoanaerobacterium Xylanolyticum With Laminaribiose
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-05-03 Classification: HYDROLASE Ligands: GOL, CA, EDO |
 |
Crystal Structure Of Unliganded Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain With A Lys329-Ile345 Loop Truncation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: EDO, SO4, NAG |
 |
Crystal Structure Of Unliganded Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: NAG, EDO |
 |
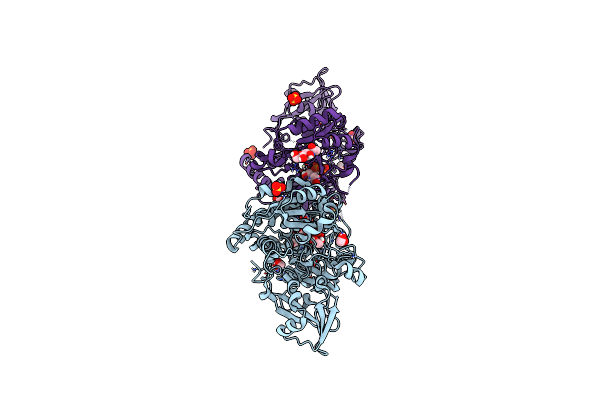
Crystal Structure Of Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain With A Lys329-Ile345 Loop Truncation, In Complex With Biantennary Pentasaccharide M592
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: EDO, NAG, SO4 |
 |
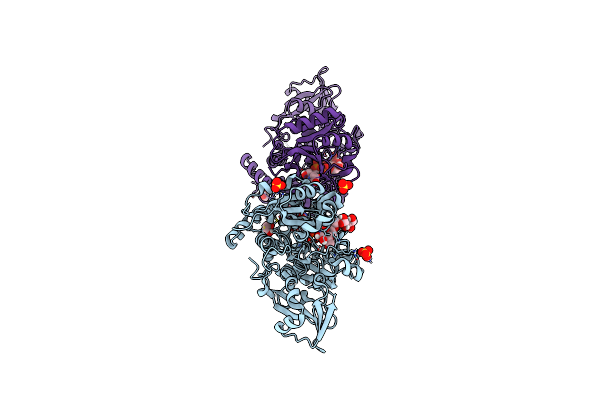
Crystal Structure Of Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain With A Lys329-Ile345 Loop Truncation, In Complex With Udp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: UDP, EDO, NAG, SO4 |
 |
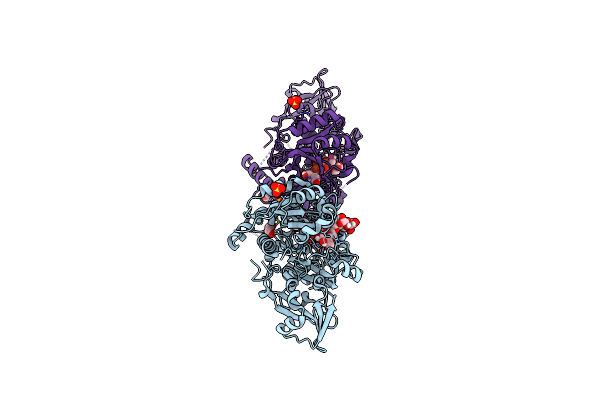
Crystal Structure Of Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain With A Lys329-Ile345 Loop Truncation, In Complex With Udp And Biantennary Pentasaccharide M592
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: UDP, EDO, SO4 |
 |
Crystal Structure Of Unliganded Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain With A Lys329-Ile345 Loop Truncation, In Complex With Udp-2-Deoxy-2-Fluoroglucose And Biantennary Pentasaccharide M592
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: EDO, U2F, SO4 |

