Search Count: 14
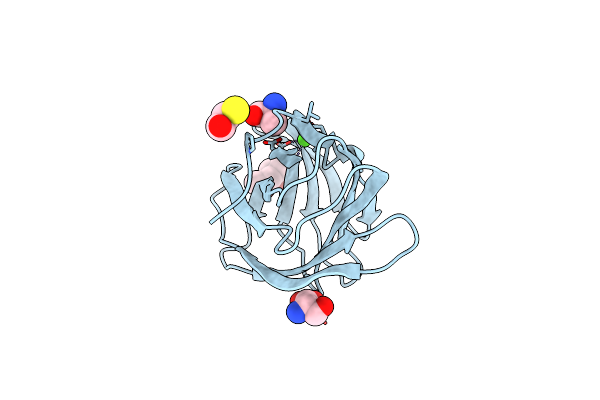 |
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-02-08 Classification: SUGAR BINDING PROTEIN Ligands: MPD, SER, BME, ALA, CA, CL |
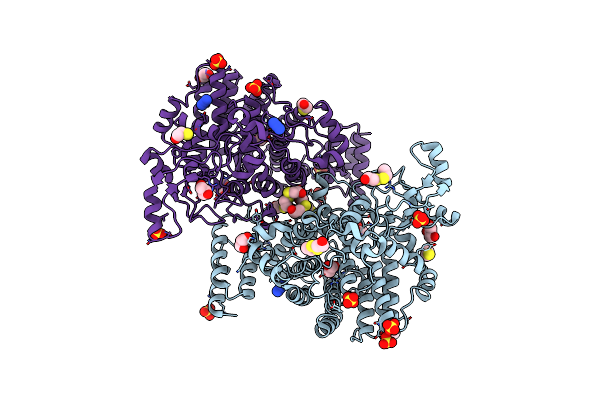 |
Organism: Bacteroides ovatus (strain atcc 8483 / dsm 1896 / jcm 5824 / nctc 11153)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-12-08 Classification: SUGAR BINDING PROTEIN Ligands: HED, BME, SO4, GOL, AZI |
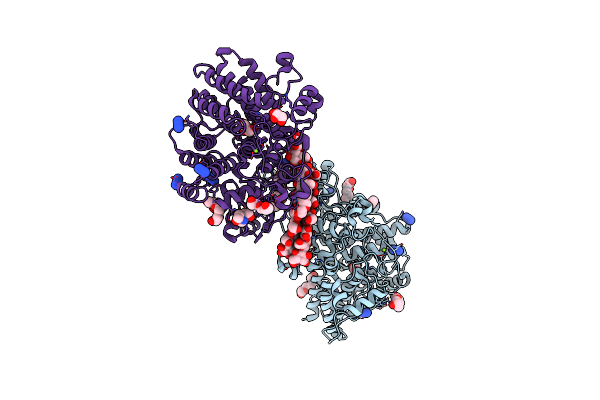 |
Structure Of Sgbp Bo2743 From Bacteroides Ovatus In Complex With Mixed-Linked Gluco-Nonasaccharide
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2021-11-10 Classification: SUGAR BINDING PROTEIN Ligands: MG, GOL, PGE, NA, AZI, TAM |
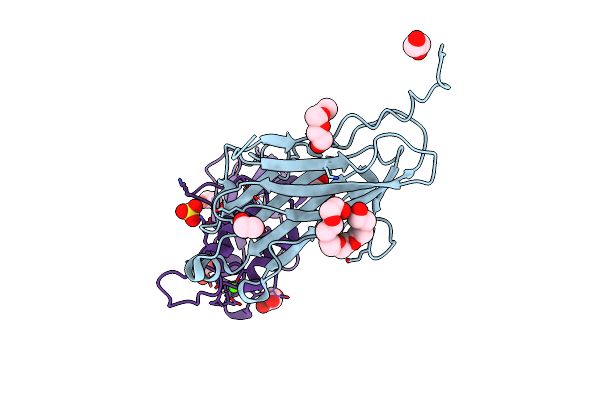 |
Crystal Structure Of The Cohscaa-Xdoccipb Type Ii Complex From Clostridium Thermocellum At 1.5Angstrom Resolution
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-09-06 Classification: CELL ADHESION Ligands: CA, SO4, GOL, EDO, PGE, P6G |
 |
Crystal Structure Of The Cohscac2-Xdoccipa Type Ii Complex From Clostridium Thermocellum
Organism: Ruminiclostridium thermocellum ad2, Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-04-05 Classification: CARBOHYDRATE BINDING PROTEIN Ligands: CA |
 |
Organism: Ruminiclostridium thermocellum 27405, Clostridium thermocellum 27405
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2017-03-29 Classification: STRUCTURAL PROTEIN Ligands: CA |
 |
High Resolution Crystallographic Structure Of The Clostridium Thermocellum N-Terminal Endo-1,4-Beta-D-Xylanase 10B (Xyn10B) Cbm22-1- Gh10 Modules Complexed With Xylohexaose
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2010-08-25 Classification: HYDROLASE Ligands: XYP, CA, PO4 |
 |
High Resolution Crystallographic Structure Of The Clostridium Thermocellum N-Terminal Endo-1,4-Beta-D-Xylanase 10B (Xyn10B) Cbm22-1- Gh10 Modules Complexed With Xylohexaose
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-08-25 Classification: HYDROLASE Ligands: CA, GOL, PO4 |
 |
High Resolution Crystallographic Structure Of The Clostridium Thermocellum N-Terminal Endo-1,4-Beta-D-Xylanase 10B (Xyn10B) Cbm22-1- Gh10 Modules Complexed With Xylohexaose
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-01-19 Classification: HYDROLASE Ligands: ACT, CD |
 |
Structure Of A Family Two Carbohydrate Esterase From Clostridium Thermocellum In Complex With Cellohexaose
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-10-06 Classification: HYDROLASE Ligands: FMT |
 |
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-03-24 Classification: HYDROLASE Ligands: ACT, GOL |
 |
Structure Of An Active Site Mutant Of A Family Two Carbohydrate Esterase From Clostridium Thermocellum In Complex With Celluohexase
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-03-24 Classification: HYDROLASE Ligands: GOL, IOD |
 |
The Clostridium Cellulolyticum Dockerin Displays A Dual Binding Mode For Its Cohesin Partner
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-05-20 Classification: CELL ADHESION Ligands: CA |
 |
The Clostridium Cellulolyticum Dockerin Displays A Dual Binding Mode For Its Cohesin Partner
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2008-05-13 Classification: CELL ADHESION Ligands: CA |

