Search Count: 31
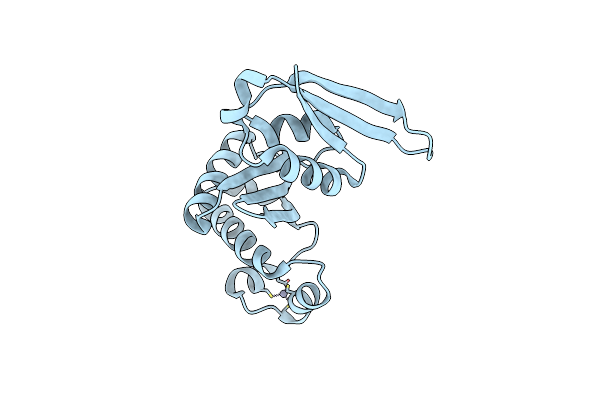 |
Organism: Saccharolobus solfataricus p2
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-10-02 Classification: RNA BINDING PROTEIN Ligands: ZN |
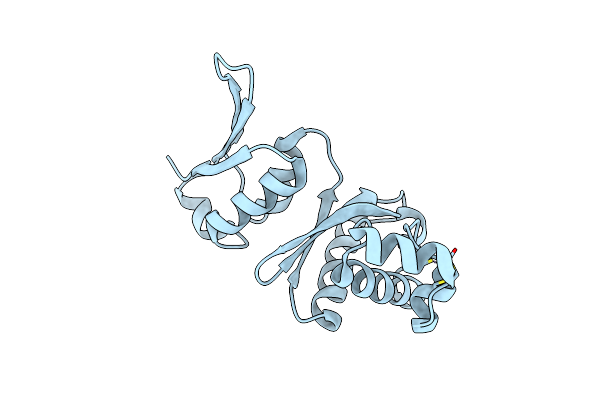 |
Organism: Sulfolobus acidocaldarius dsm 639
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-09-11 Classification: UNKNOWN FUNCTION Ligands: ZN |
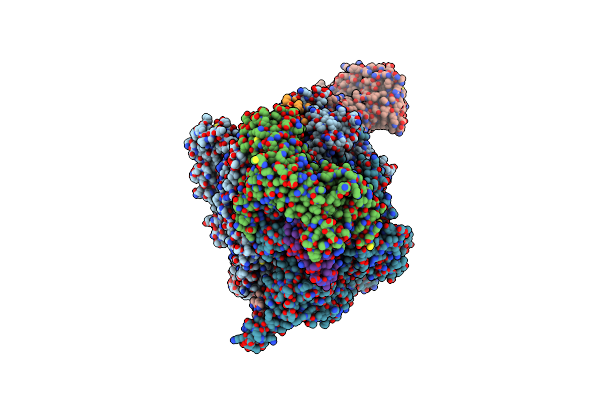 |
Organism: African swine fever virus ba71v
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSCRIPTION Ligands: ZN, MG |
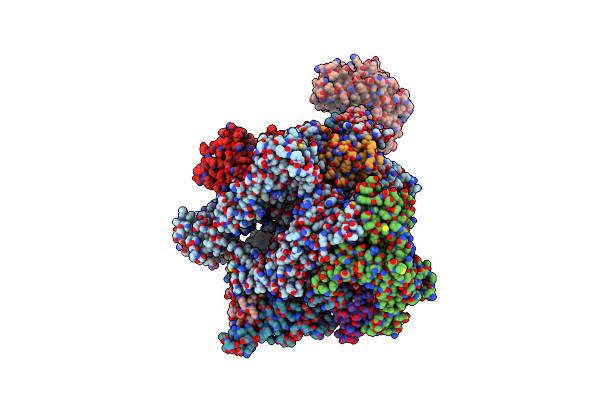 |
Organism: African swine fever virus ba71v
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Cryo-Em Structure Of The Sulfolobus Acidocaldarius Rna Polymerase At 2.88 A
Organism: Sulfolobus acidocaldarius (strain atcc 33909 / dsm 639 / jcm 8929 / nbrc 15157 / ncimb 11770)
Method: ELECTRON MICROSCOPY Release Date: 2021-08-25 Classification: TRANSCRIPTION Ligands: MG, ZN, F3S |
 |
Cryo-Em Structure Of The Atv Rnap Inhibitory Protein (Rip) Bound To The Dna-Binding Channel Of The Host'S Rna Polymerase
Organism: Sulfolobus acidocaldarius dsm 639, Acidianus two-tailed virus
Method: ELECTRON MICROSCOPY Release Date: 2021-08-25 Classification: VIRAL PROTEIN Ligands: MG, ZN, F3S |
 |
Cryo-Em Structure Of The Cellular Negative Regulator Tfs4 Bound To The Archaeal Rna Polymerase
Organism: Sulfolobus acidocaldarius dsm 639, Saccharolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2)
Method: ELECTRON MICROSCOPY Release Date: 2021-08-25 Classification: TRANSCRIPTION Ligands: MG, ZN, F3S |
 |
Structure Of Lsd2/Npac-Linker/Nucleosome Core Particle Complex: Class 1, Free Nuclesome
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:4.02 Å Release Date: 2019-04-24 Classification: GENE REGULATION |
 |
Organism: Xenopus laevis, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2019-04-24 Classification: GENE REGULATION Ligands: FAD, ZN |
 |
Organism: Homo sapiens, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2019-04-24 Classification: GENE REGULATION Ligands: FAD, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-09-21 Classification: OXIDOREDUCTASE/REPRESSOR Ligands: FAD, E11, NA |
 |
Organism: Homo sapiens, Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-09-21 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Homo sapiens, Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-09-21 Classification: OXIDOREDUCTASE/REPRESSOR Ligands: FAD, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2016-09-21 Classification: OXIDOREDUCTASE/REPRESSOR Ligands: FAD, 767 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2016-05-04 Classification: OXIDOREDUCTASE/REPRESSOR Ligands: FAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.31 Å Release Date: 2016-05-04 Classification: OXIDOREDUCTASE/REPRESSOR Ligands: FAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-05-04 Classification: OXIDOREDUCTASE/REPRESSOR Ligands: FAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-11-27 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2013-06-12 Classification: OXIDOREDUCTASE/PEPTIDE Ligands: FAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-06-12 Classification: OXIDOREDUCTASE/PEPTIDE Ligands: FAD |

