Search Count: 20
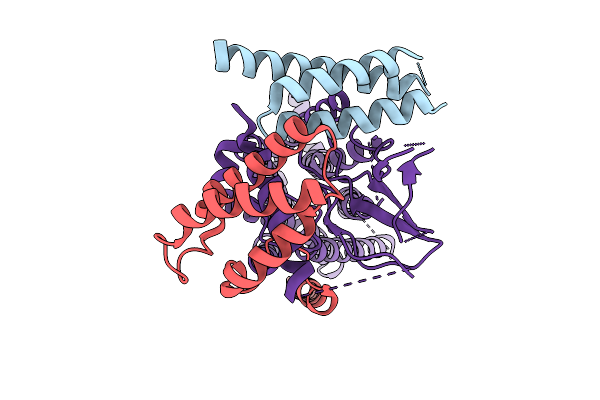 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN |
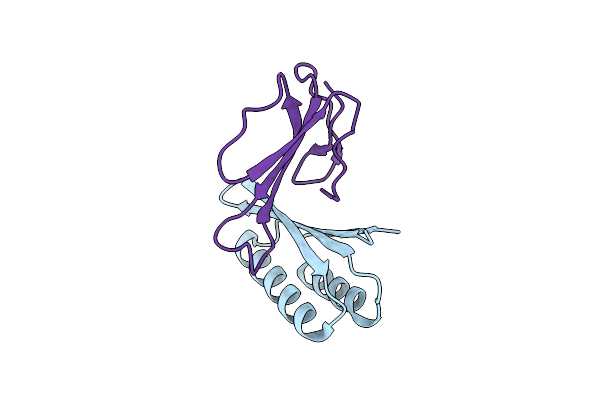 |
Organism: Synthetic construct, Naja kaouthia
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2024-10-09 Classification: DE NOVO PROTEIN/TOXIN |
 |
Organism: Synthetic construct, Naja pallida
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-09 Classification: DE NOVO PROTEIN/TOXIN |
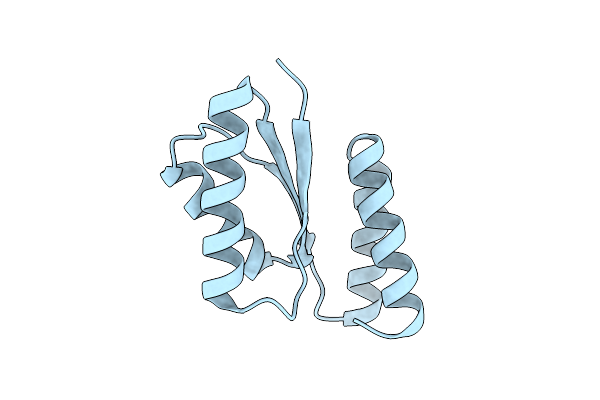 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-10-09 Classification: DE NOVO PROTEIN |
 |
Cryoem Structure Of Allosterically Switchable De Novo Protein Sr322, In Closed State Without Effector Peptide
Organism: Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: DE NOVO PROTEIN |
 |
Cryoem Structure Of Allosterically Switchable De Novo Protein Sr312, In Open State With Effector Peptide
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: DE NOVO PROTEIN |
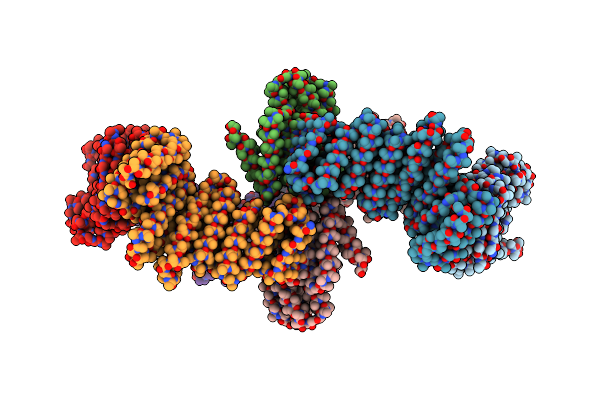 |
Cryoem Structure Of Allosterically Switchable De Novo Protein Sr322, In Closed State Without Effector Peptide, Off Target Multimeric State
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: DE NOVO PROTEIN |
 |
Organism: Methylophaga sulfidovorans
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-07-31 Classification: TRANSFERASE |
 |
Organism: Ananas comosus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSFERASE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2024-07-17 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.59 Å Release Date: 2024-07-17 Classification: DE NOVO PROTEIN Ligands: CHD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: MEMBRANE PROTEIN Ligands: PEF, NAG, CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: MEMBRANE PROTEIN Ligands: NAG, CA, AQV |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-08-16 Classification: DE NOVO PROTEIN Ligands: PO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-08-16 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2023-08-16 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-08-16 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2023-08-16 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-07-27 Classification: RNA BINDING PROTEIN Ligands: MGO |
 |
Engineering Disulphide-Free Autonomous Antibody Vh Domains To Modulate Intracellular Pathways
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2021-08-25 Classification: RNA BINDING PROTEIN |

