Search Count: 65
 |
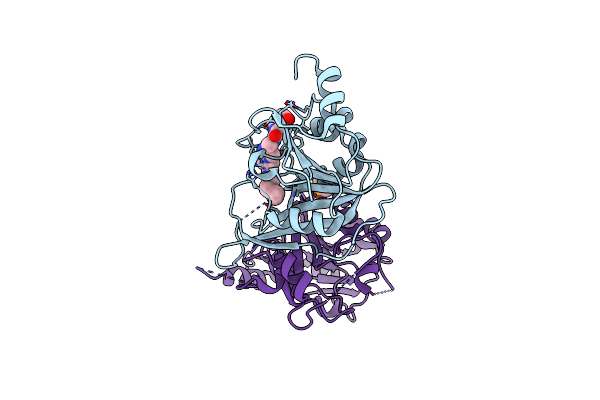
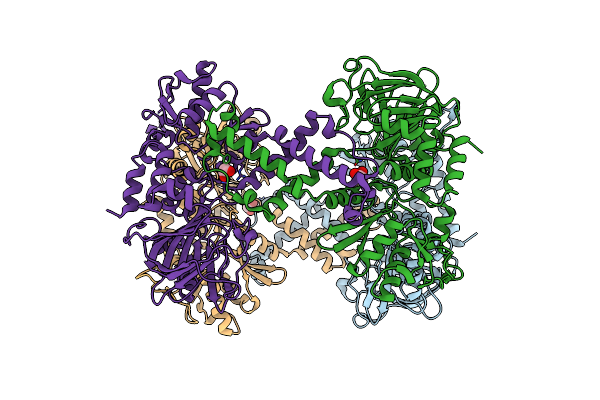
Mettl3-Mettl14 Heterodimer Bound To The Sam Competitive Small Molecule Inhibitor Stm3006
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2023-09-20 Classification: RNA BINDING PROTEIN Ligands: QWR |
 |
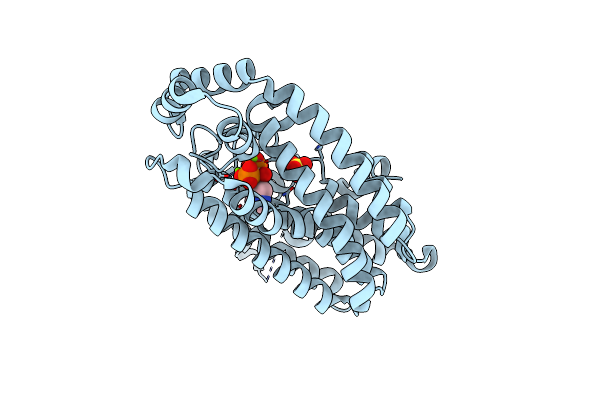
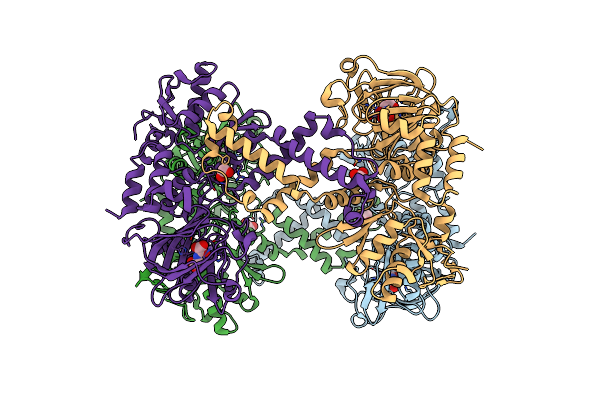
Mettl3-Mettl14 Heterodimer Bound To The Sam Competitive Small Molecule Inhibitor Stm2457
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-04-14 Classification: RNA BINDING PROTEIN Ligands: V22, DMS |
 |
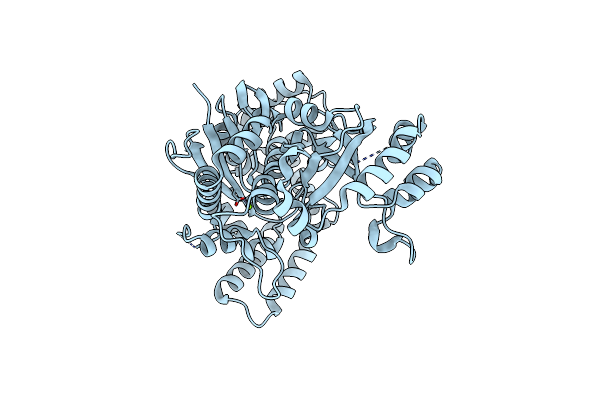
The Role Of Threonine 201 And Tyrosine 204 In The Human Farnesyl Pyrophosphate Synthase Catalytic Mechanism And The Mode Of Inhibition By The Nitrogen-Containing Bisphosphonates
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2015-03-18 Classification: TRANSFERASE Ligands: RIS, MG, SO4 |
 |
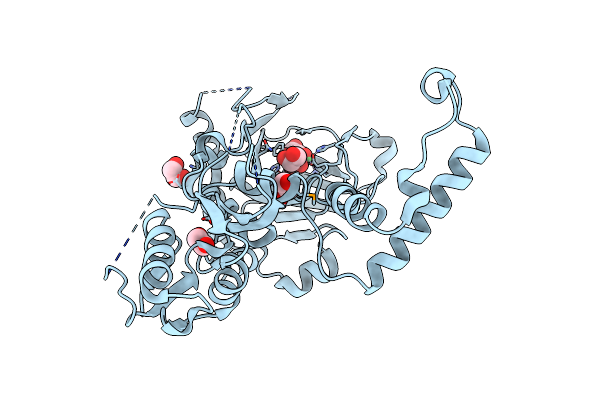
Structural And Biochemical Studies Of A Moderately Thermophilic Exonuclease I From Methylocaldum Szegediense
Organism: Methylocaldum szegediense
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2015-02-25 Classification: HYDROLASE Ligands: MG |
 |
60S Ribosomal Protein L27A Histidine Hydroxylase (Mina53) In Complex With Ni(Ii) And 2-Oxoglutarate (2Og)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: NI, AKG, EDO |
 |
60S Ribosomal Protein L27A Histidine Hydroxylase (Mina53 Y209C) In Complex With Mn(Ii), 2-Oxoglutarate (2Og) And 60S Ribosomal Protein L27A (Rpl27A G37C) Peptide Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE/TRANSLATION Ligands: MN, AKG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: EDO |
 |
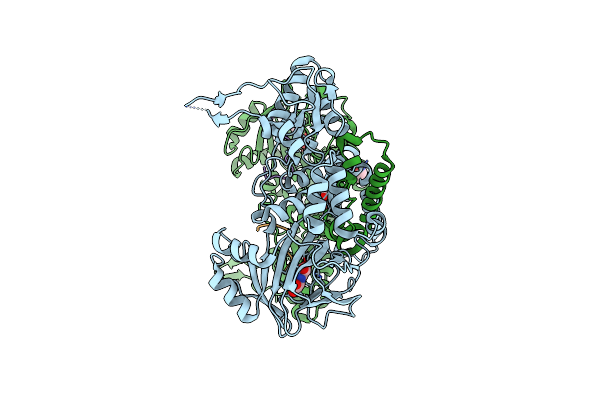
60S Ribosomal Protein L8 Histidine Hydroxylase (No66) In Complex With Mn(Ii) And N-Oxalylglycine (Nog)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: MN, OGA, EDO |
 |
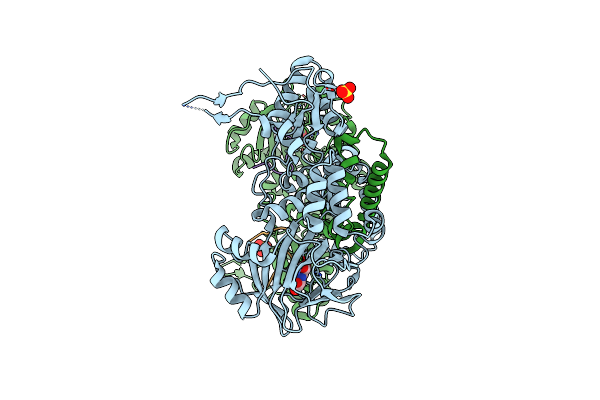
60S Ribosomal Protein L8 Histidine Hydroxylase (No66) In Complex With Mn(Ii), N-Oxalylglycine (Nog) And 60S Ribosomal Protein L8 (Rpl8 G220C) Peptide Fragment (Complex-1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: MN, OGA, EDO |
 |
60S Ribosomal Protein L8 Histidine Hydroxylase (No66 L299C/C300S) In Complex With Mn(Ii), N-Oxalylglycine (Nog) And 60S Ribosomal Protein L8 (Rpl8 G220C) Peptide Fragment (Complex-2)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: MN, OGA, SO4, EDO |
 |
60S Ribosomal Protein L8 Histidine Hydroxylase (No66 S373C) In Complex With Mn(Ii), N-Oxalylglycine (Nog) And 60S Ribosomal Protein L8 (Rpl8 G214C) Peptide Fragment (Complex-3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: MN, OGA, EDO |
 |
Structure Of Ycfd A Ribosomal Oxygenase From Escherichia Coli In Complex With Cobalt And 2-Oxoglutarate.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: CO, AKG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: TAM, PO4 |
 |
Structure Of Ycfd, A Ribosomal Oxygenase From Escherichia Coli In Complex With Cobalt And Succinic Acid.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: CO, SIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-04-30 Classification: OXIDOREDUCTASE Ligands: MN, SO4 |
 |
Rhodothermus Marinus Ycfd-Like Ribosomal Protein L16 Arginyl Hydroxylase In Complex Substrate Fragment
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2014-04-16 Classification: TRANSLATION Ligands: MN, OGA, MPD, PO4 |
 |
Organism: Rhodothermus marinus
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2014-04-09 Classification: OXIDOREDUCTASE Ligands: CIT, UN9, GOL, MN |
 |
Crystal Structure Of Malonyl-Coa Decarboxylase (Rmet_2797) From Cupriavidus Metallidurans, Northeast Structural Genomics Consortium Target Crr76
Organism: Cupriavidus metallidurans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-06-19 Classification: LYASE Ligands: MG |
 |
Crystal Structure Of Malonyl-Coa Decarboxylase From Rhodopseudomonas Palustris, Northeast Structural Genomics Consortium Target Rpr127
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-06-19 Classification: LYASE Ligands: MG |
 |
Crystal Structure Of Malonyl-Coa Decarboxylase From Agrobacterium Vitis, Northeast Structural Genomics Consortium Target Rir35
Organism: Agrobacterium vitis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-06-19 Classification: LYASE Ligands: NI, CL |

