Search Count: 32
 |
Crystal Structure Of A Selenomethionine-Substituted A70M I84M Mutant Of The Essential Repressor Ddro From Radiation Resistant-Deinococcus Bacteria (Deinococcus Deserti)
Organism: Deinococcus deserti
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-10-09 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of The Essential Repressor Ddro From Radiation-Resistant Deinococcus Bacteria (Deinococcus Deserti)
Organism: Deinococcus deserti (strain vcd115 / dsm 17065 / lmg 22923)
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2019-10-09 Classification: DNA BINDING PROTEIN Ligands: CL |
 |
Crystal Structure Of The N-Terminal Hth Dna-Binding Domain Of The Essential Repressor Ddro From Radiation-Resistant Deinococcus Bacteria (Deinococcus Deserti)
Organism: Deinococcus deserti
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2019-10-09 Classification: DNA BINDING PROTEIN Ligands: GOL |
 |
Crystal Structure Of The C-Terminal Dimerization Domain Of The Essential Repressor Ddro From Radiation-Resistant Deinococcus Bacteria (Deinococcus Deserti)
Organism: Deinococcus deserti
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2019-10-09 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2019-04-10 Classification: BIOSYNTHETIC PROTEIN Ligands: NH4, CIT, PG4 |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-04-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 2PE, GOL |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-04-10 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, DHI, GOL, PG4 |
 |
Organism: Staphylococcus aureus subsp. aureus n315
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2019-04-10 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP, 8UX, GOL, PG4 |
 |
Structure Of Fatty Acid Photodecarboxylase In Complex With Fad And Palmitic Acid
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2017-08-30 Classification: OXIDOREDUCTASE Ligands: FAD, PLM |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-05-20 Classification: HYDROLASE Ligands: GDP |
 |
Organism: Magnetococcus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-10-09 Classification: ELECTRON TRANSPORT Ligands: HEC, GOL |
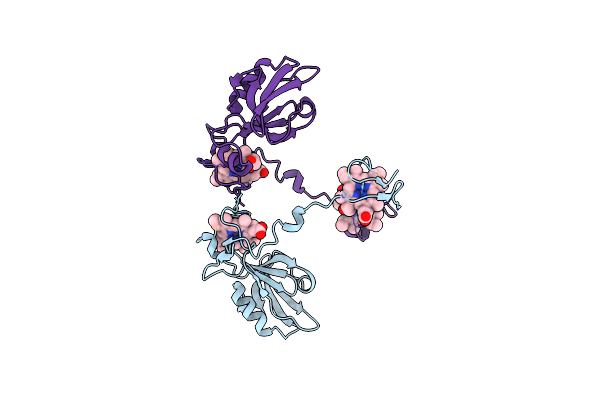 |
Organism: Magnetococcus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-10-09 Classification: ELECTRON TRANSPORT Ligands: HEC |
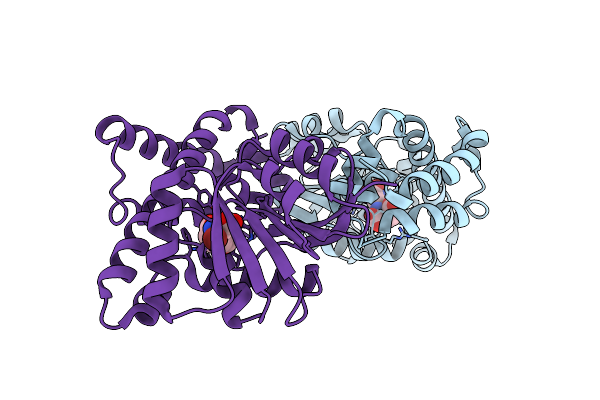 |
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-06-08 Classification: BIOSYNTHETIC PROTEIN Ligands: 3O3, BR |
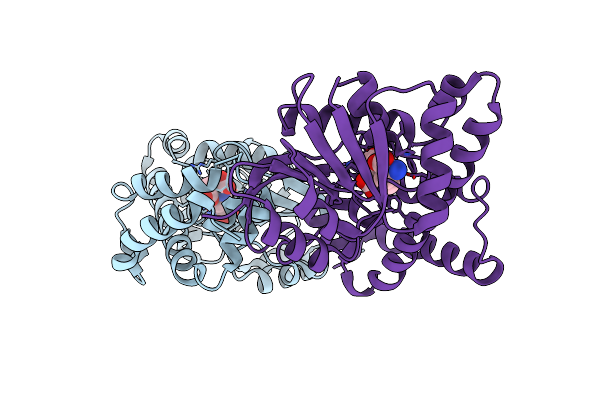 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-10-06 Classification: BIOSYNTHETIC PROTEIN, TRANSFERASE Ligands: TNA, BR |
 |
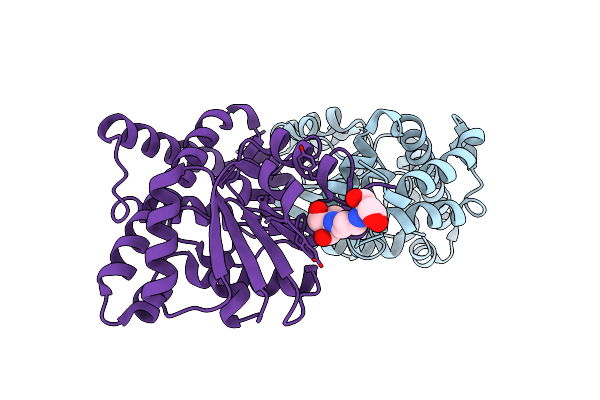
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2009-10-06 Classification: BIOSYNTHETIC PROTEIN, TRANSFERASE Ligands: TNA, MTA, BR |
 |
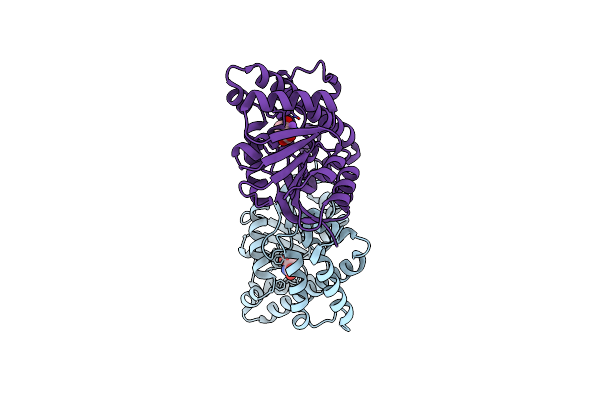
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-10-06 Classification: BIOSYNTHETIC PROTEIN, TRANSFERASE Ligands: BR, B3P |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-10-06 Classification: BIOSYNTHETIC PROTEIN, TRANSFERASE Ligands: GLU, NA |
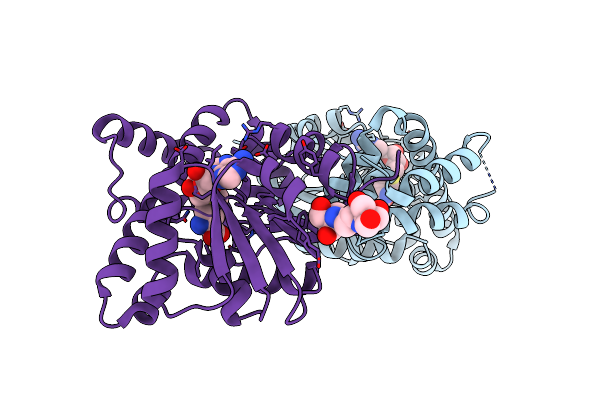 |
Crystal Structure Of E81Q Mutant Of Mtnas In Complex With S-Adenosylmethionine
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-10-06 Classification: BIOSYNTHETIC PROTEIN, TRANSFERASE Ligands: SAM, BR, B3P |
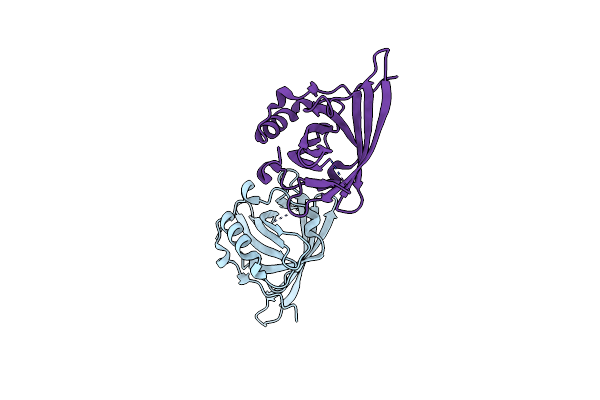 |
Crystal Structure Of The Lipocalin Domain Of Violaxanthin De-Epoxidase (Vde) At Ph7
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-04-21 Classification: OXIDOREDUCTASE |
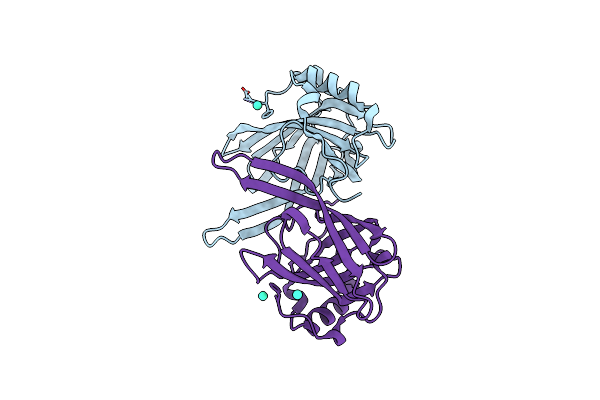 |
Crystal Structure Of The Lipocalin Domain Of Violaxanthin De-Epoxidase (Vde) At Ph5
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-04-21 Classification: OXIDOREDUCTASE Ligands: GD |

