Search Count: 28
 |
Organism: Rhodospirillum rubrum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: OXIDOREDUCTASE Ligands: S5Q, CLF, ADP, MG, AF3, SF4 |
 |
Organism: Rhodobacter capsulatus sb 1003
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: ELECTRON TRANSPORT Ligands: FES |
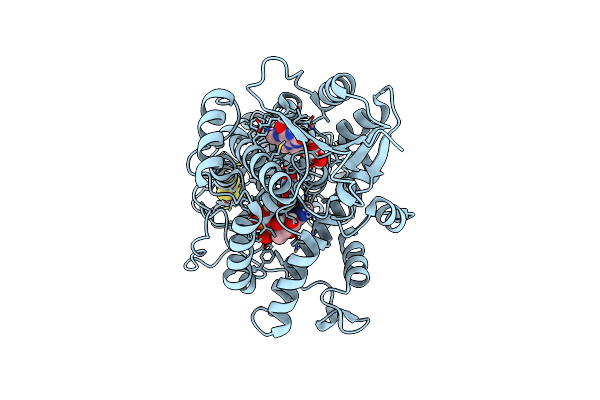 |
Structure Of The (6-4) Photolyase Of Caulobacter Crescentus In Its Oxidizd State - Singal Crystal / Synchrotron
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DLZ, SF4 |
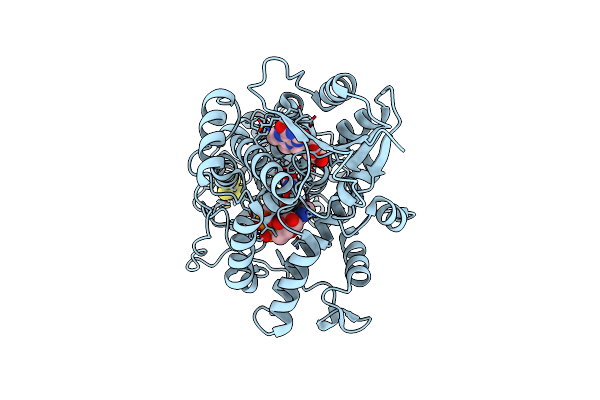 |
Structure Of The (6-4) Photolyase Of Caulobacter Crescentus In Its Oxidized State At Room Temperture-Synchrotron
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DLZ, SF4 |
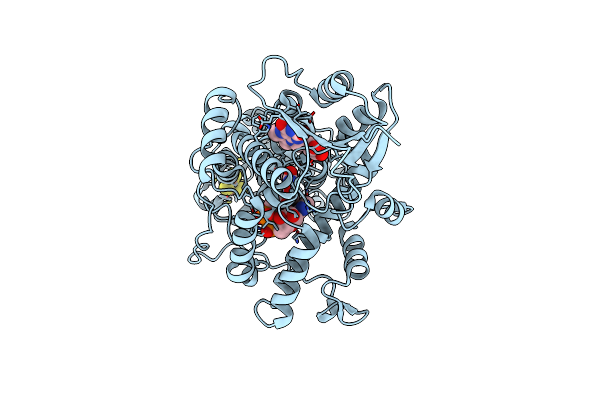 |
Structure Of The (6-4) Photolyase Of Caulobacter Crescentus In Its Dark Adapted And Oxidized State Determined By Serial Femtosecond Crystallography
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DLZ, SF4 |
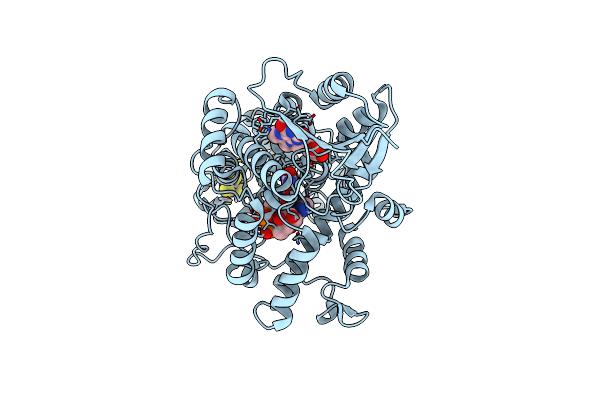 |
Structure Of The (6-4) Photolyase Of Caulobacter Crescentus In Its Fully Reduced State Determined By Serial Femtosecond Crystallography
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DLZ, SF4 |
 |
Structure Of The (6-4) Photolyase Of Caulobacter Crescentus In Its Iron Sulfur Cluster Oxidized State Determined By Serial Femtosecond Crystallography
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DLZ, SF4 |
 |
Structure Of The (6-4) Photolyase Of Caulobacter Crescentus With K48A Mutation In Its Dark Adapted And Oxidized State Determined By Synchrotron
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DLZ, SF4, GOL, CL, SO4, MG |
 |
Organism: Azoarcus sp. cib
Method: X-RAY DIFFRACTION Release Date: 2025-03-05 Classification: OXIDOREDUCTASE Ligands: BYC, GOL, SF4, BJ8, TRS, DOD |
 |
Organism: Azoarcus sp. cib
Method: X-RAY DIFFRACTION Release Date: 2025-03-05 Classification: OXIDOREDUCTASE Ligands: 4KX, TRS, GOL, SF4, BJ8 |
 |
Organism: Azoarcus sp. cib
Method: X-RAY DIFFRACTION Release Date: 2025-03-05 Classification: OXIDOREDUCTASE Ligands: BYC, GOL, SF4, BJ8, TRS, DOD |
 |
Crystal Structure Of The Coenzyme F420-Dependent Sulfite Reductase From Methanocaldococcus Jannaschii At 2.3-A Resolution
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-03-23 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, SO3, SRM, GOL, MPD, CA, CL, FE |
 |
Crystal Structure Of The Coenzyme F420-Dependent Sulfite Reductase From Methanothermococcus Thermolithotrophicus At 1.55-A Resolution
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-03-23 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, EDO, SRM, SO4, H2S, CL, GOL, LI, PEG, TRS |
 |
Organism: Bacterium enrichment culture clone n47
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-05-15 Classification: FLAVOPROTEIN Ligands: SF4, FAD, FMN |
 |
Organism: Bacterium enrichment culture clone n47
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-05-15 Classification: FLAVOPROTEIN Ligands: SF4, FAD, FMN, J5H |
 |
2-Naphthoyl-Coa Reductase-Dihydronaphthoyl-Coa Complex(Ncr-Dhncoa Co-Crystallized Complex)
Organism: Bacterium enrichment culture clone n47
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-05-15 Classification: FLAVOPROTEIN Ligands: SF4, FAD, FMN, J5H |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-01-14 Classification: TRANSLATION Ligands: FE, SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-01-14 Classification: TRANSLATION Ligands: FE, SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-01-14 Classification: TRANSLATION |
 |
The F420-Reducing [Nife]-Hydrogenase Complex From Methanothermobacter Marburgensis, The First X-Ray Structure Of A Group 3 Family Member
Organism: Methanothermobacter marburgensis
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2014-06-11 Classification: OXIDOREDUCTASE Ligands: SF4, DTZ, UNL, MG, NFU, CL, KEN, FAD |

