Search Count: 37
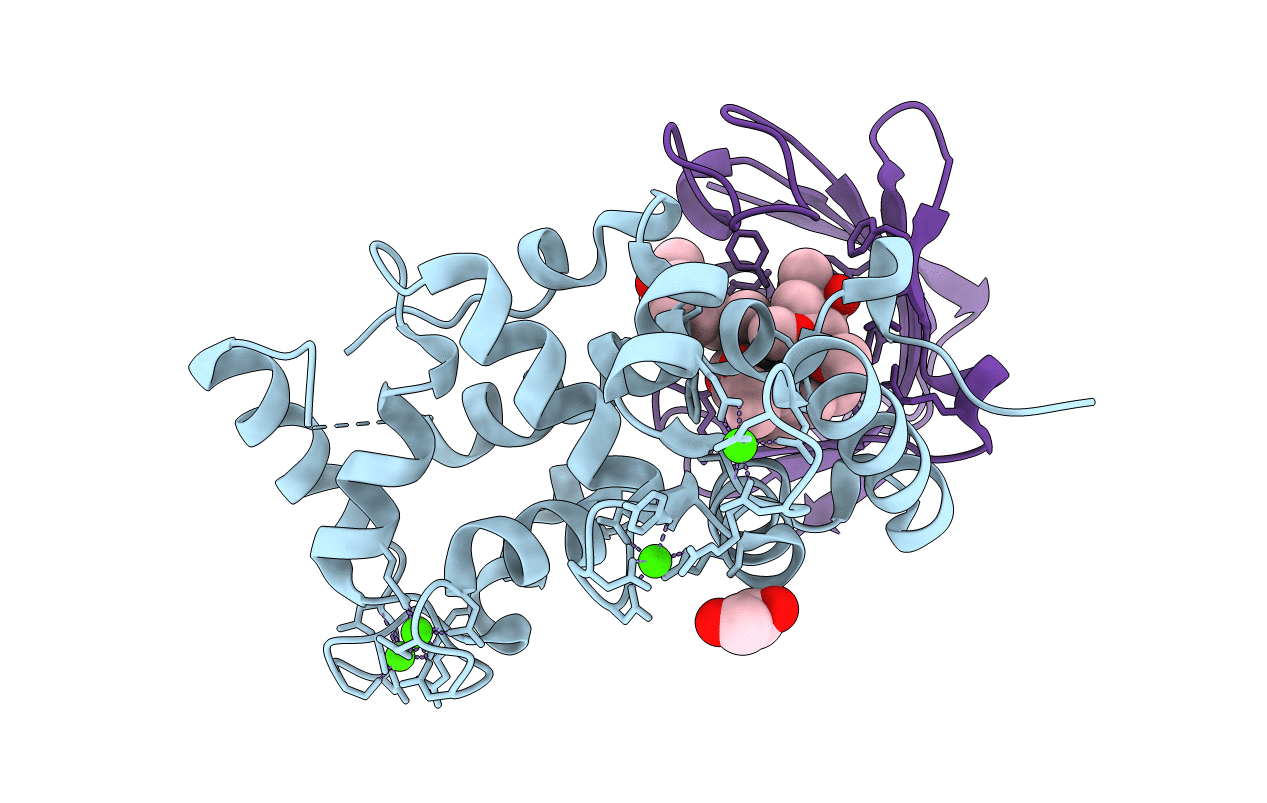 |
Crystal Structure Of A Human Calcineurin A - Calcineurin B Fusion Bound To Fkbp12 And Fk-520
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-08-03 Classification: HYDROLASE/ISOMERASE Ligands: CA, EDO, KXX |
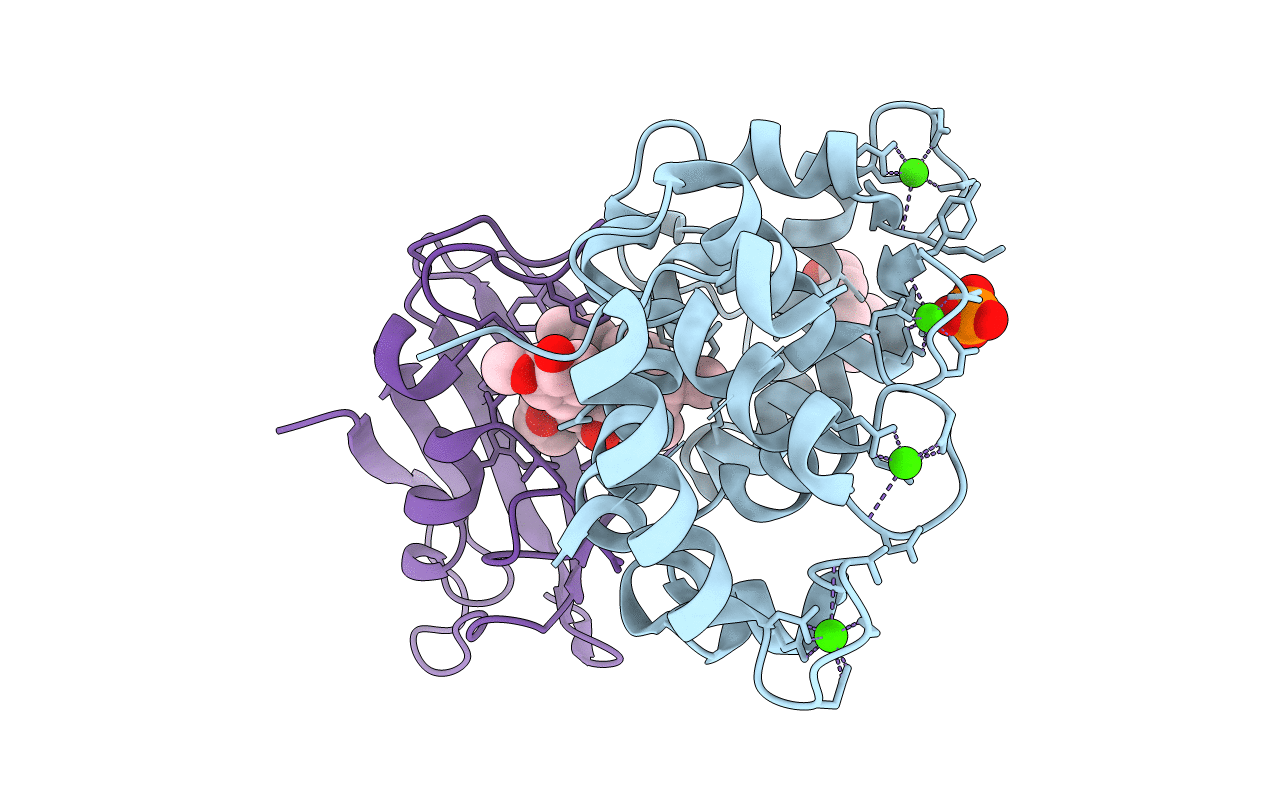 |
Crystal Structure Of A Aspergillus Fumigatus Calcineurin A - Calcineurin B Fusion Bound To Fkbp12 And Fk-506
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-08-03 Classification: ANTIFUNGAL PROTEIN Ligands: CA, PG4, PO4, FK5 |
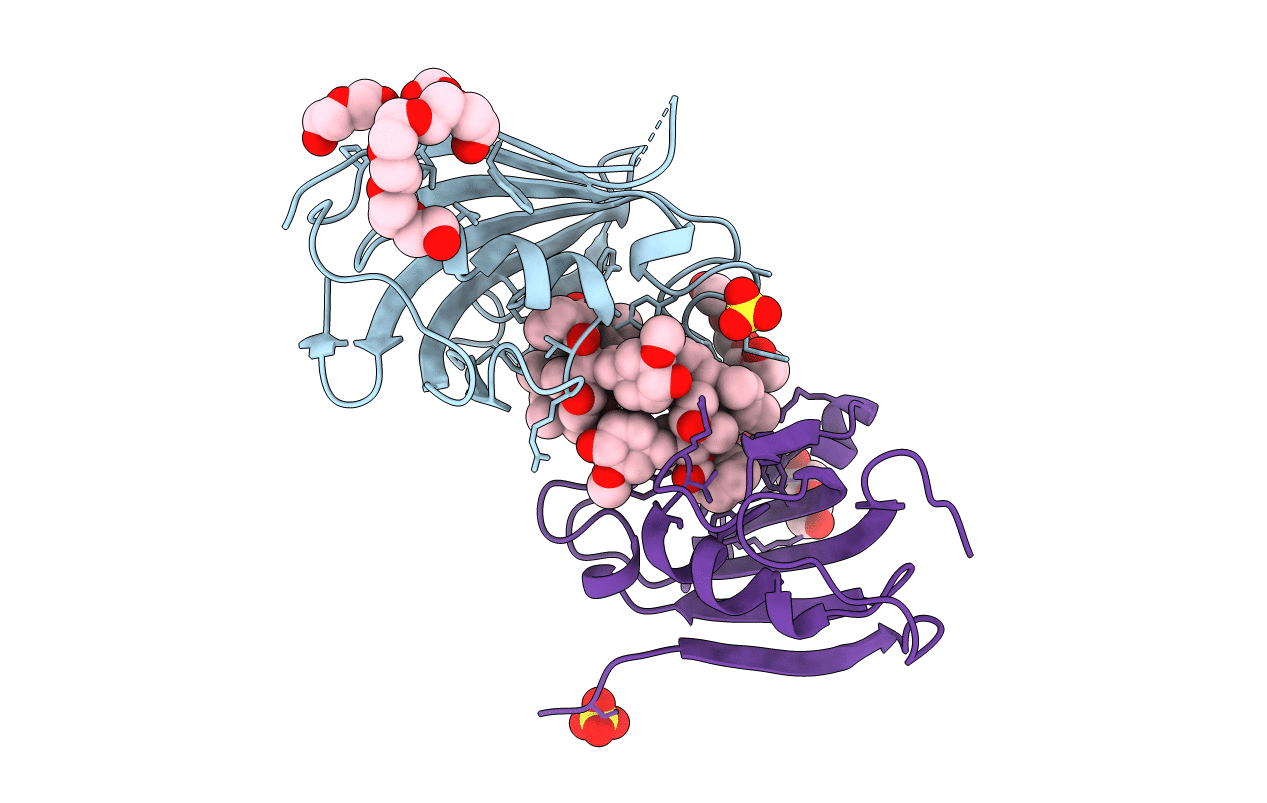 |
Crystal Structure Of Fk506-Binding Protein 1A From Aspergillus Fumigatus Bound To Ascomycin
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-07-27 Classification: ANTIFUNGAL PROTEIN Ligands: KXX, SO4, PG4, P6G, ACT, 1PE |
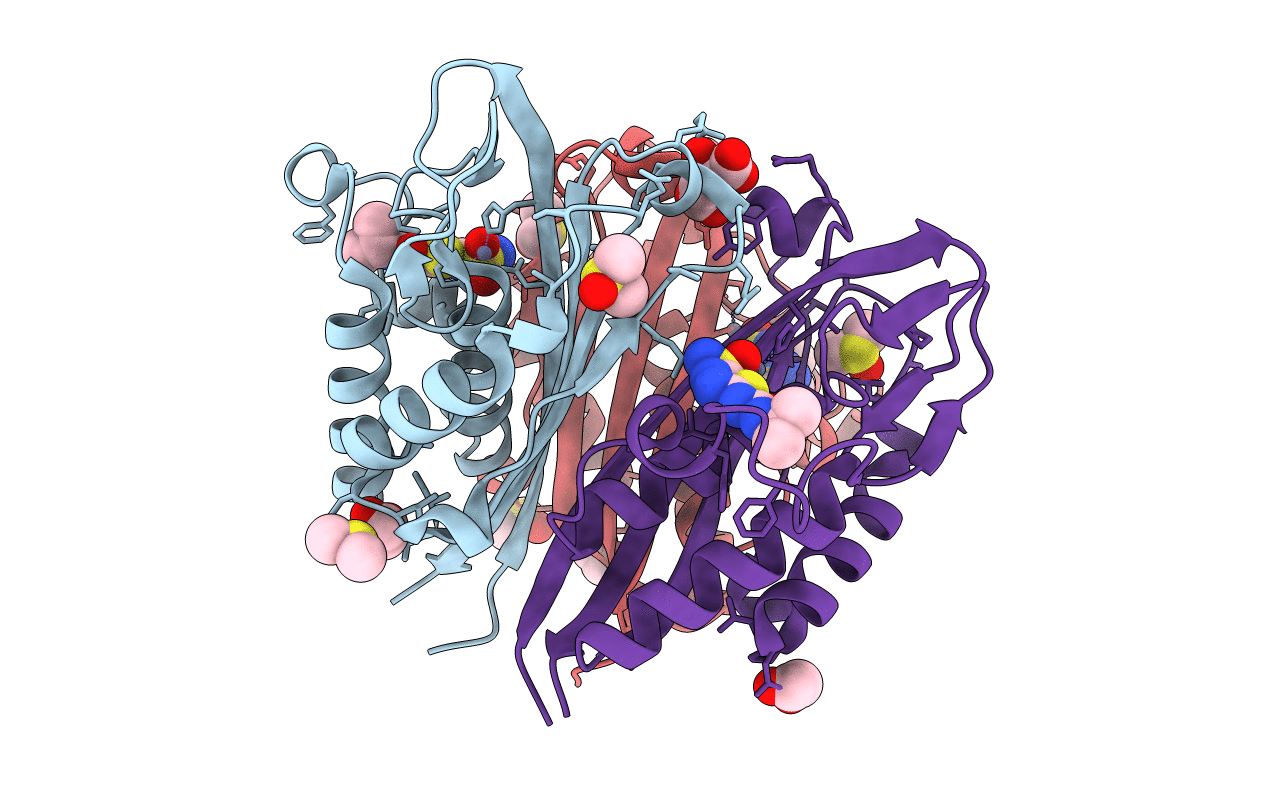 |
Crystal Structure Of 2C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase (Ispf) Burkholderia Pseudomallei In Compomplex With Ligand Hgn-0961 (Bsi110840)
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-12-09 Classification: LYASE Ligands: ZN, QOG, DMS, EDO, MLT |
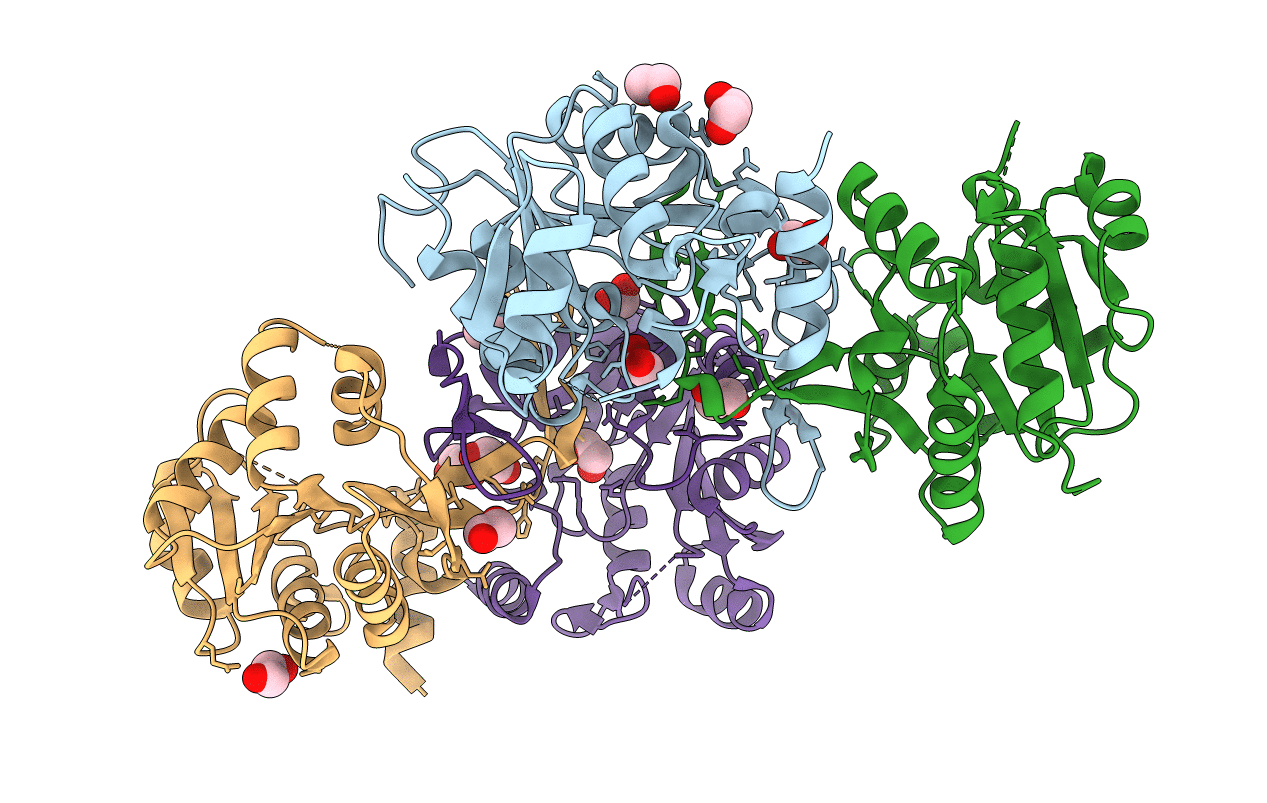 |
Organism: Mycolicibacterium paratuberculosis (strain atcc baa-968 / k-10)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-11-11 Classification: TRANSFERASE Ligands: EDO |
 |
Crystal Structure Of Smt Fusion Peptidyl-Prolyl Cis-Trans Isomerase From Burkholderia Pseudomallei Complexed With Sf339
Organism: Saccharomyces cerevisiae, Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-02-12 Classification: ISOMERASE, PROTEIN BINDING Ligands: LL7, CA, EDO |
 |
Crystal Structure Of Smt Fusion Peptidyl-Prolyl Cis-Trans Isomerase From Burkholderia Pseudomallei Complexed With Sf355
Organism: Saccharomyces cerevisiae, Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-02-12 Classification: ISOMERASE, PROTEIN BINDING Ligands: CA, LLD |
 |
Crystal Structure Of 2C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase (Ispf) Burkholderia Pseudomallei In Complex With Ligand Sr-4
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-01-15 Classification: LYASE Ligands: ZN, DMS, MLI, EDO, QMS |
 |
Crystal Structure Of 2C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase (Ispf) Burkholderia Pseudomallei In Complex With Ligand Hgn-0459
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-11-06 Classification: LYASE Ligands: ZN, SFY, DMS, ACT |
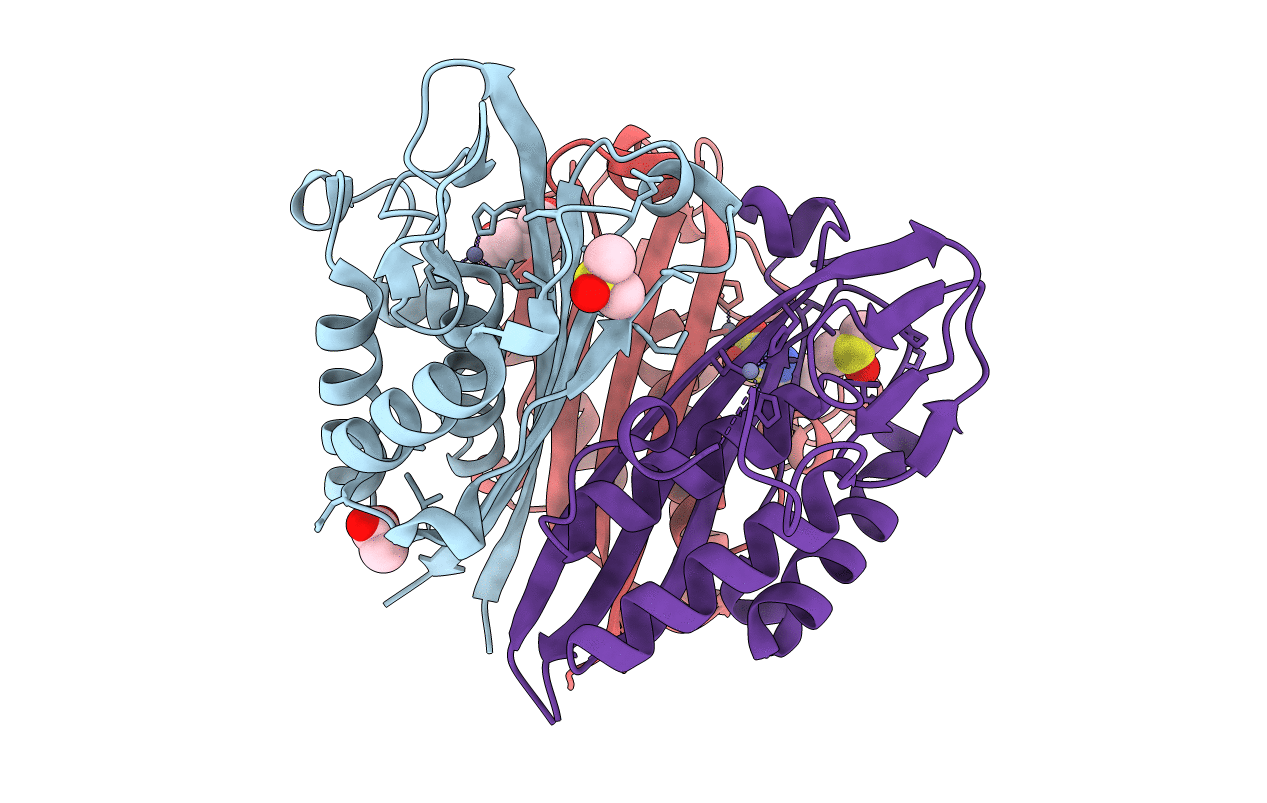 |
Crystal Structure Of 2C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase (Ispf) Burkholderia Pseudomallei In Complex With Ligand Hgn-0456
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-11-06 Classification: LYASE Ligands: ZN, DMS, ACT, EZL, PEG |
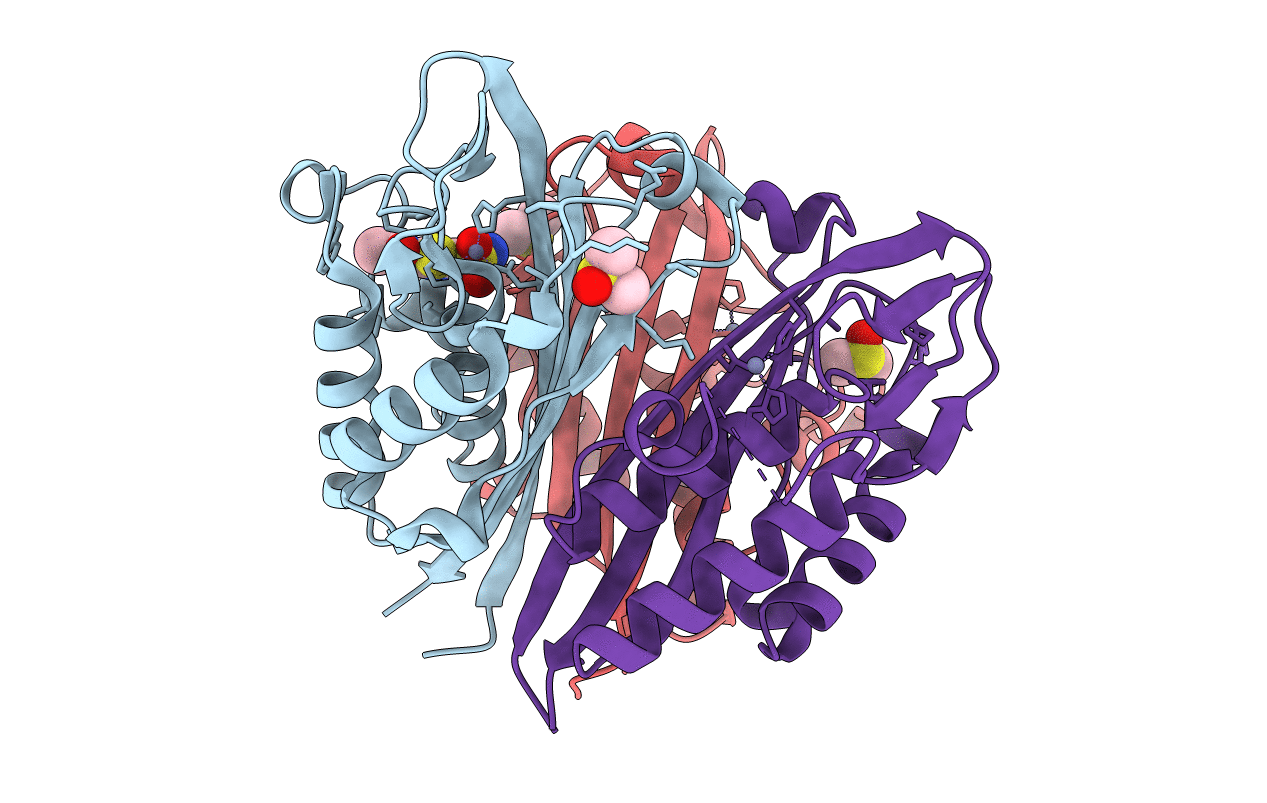 |
Crystal Structure Of 2C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase (Ispf) Burkholderia Pseudomallei In Complex With Ligand Hgn-0863
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-11-06 Classification: LYASE Ligands: ZN, AZM, DMS |
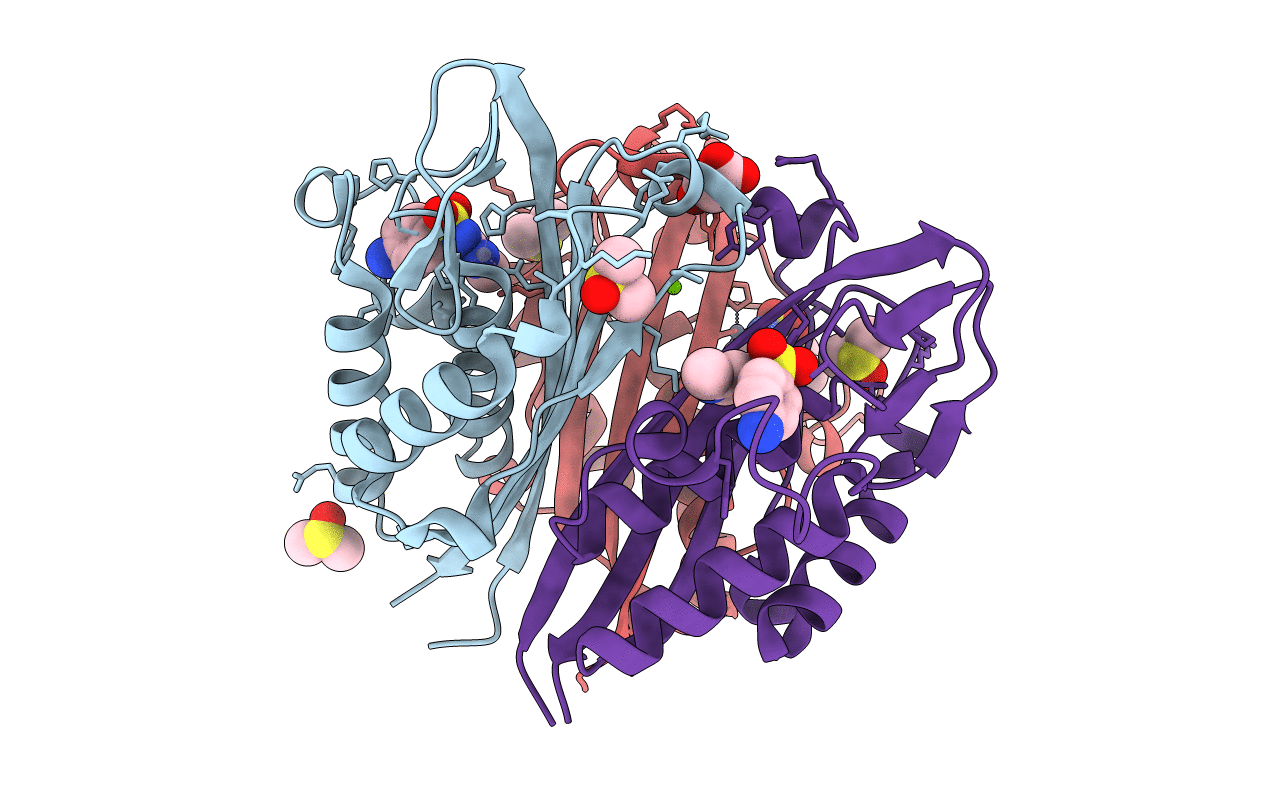 |
Crystal Structure Of 2C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase (Ispf) Burkholderia Pseudomallei In Complex With Ligand Hgn-0883
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-11-06 Classification: LYASE Ligands: ZN, K4Y, DMS, MG, MLA |
 |
Crystal Structure Of 3-Ketoacyl-(Acyl-Carrier-Protein) Reductase From Brucella Melitensis Complexed With Nadh
Organism: Brucella abortus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-10-25 Classification: OXIDOREDUCTASE Ligands: NAI |
 |
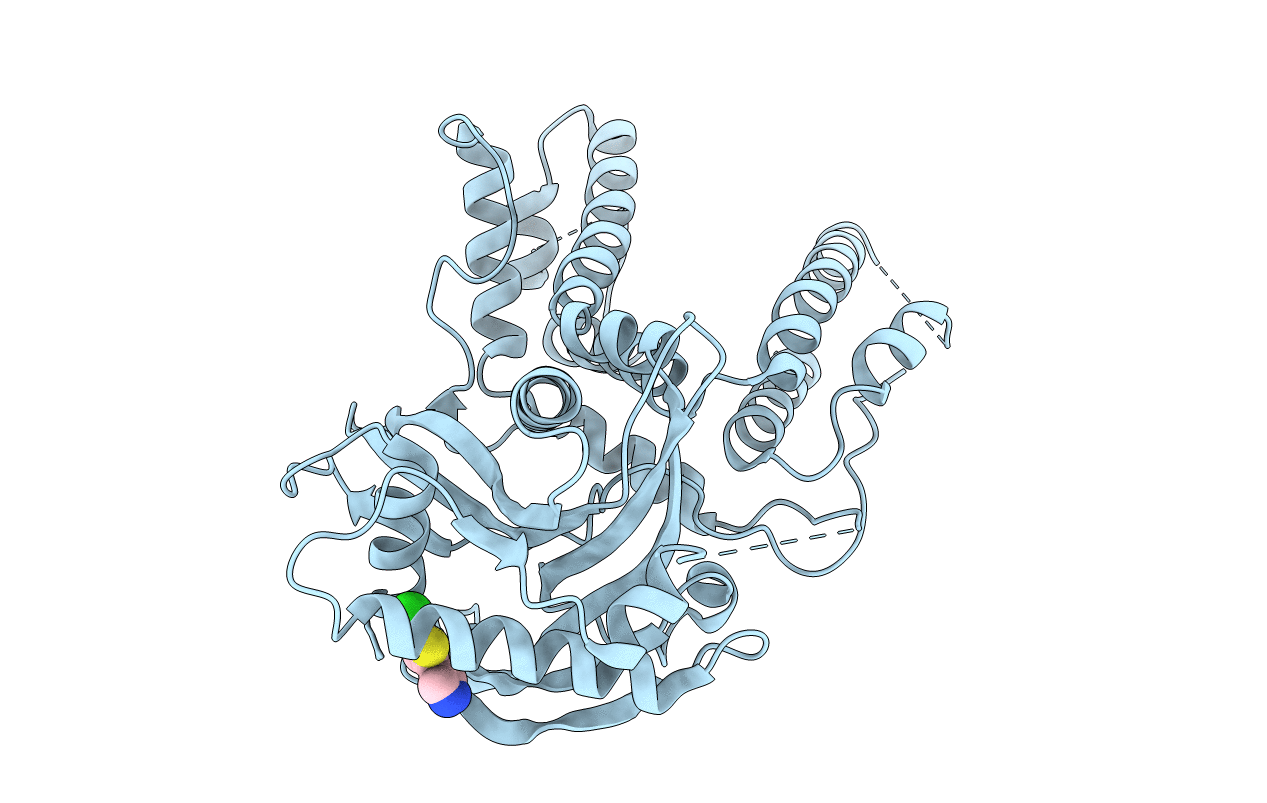
Crystal Structure Of The C-Terminal Domain Of Doublecortin (Tgdcx) From Toxoplasma Gondii Me49
Organism: Toxoplasma gondii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-10-18 Classification: STRUCTURAL PROTEIN Ligands: FMT |
 |
Crystal Structure Of Polymerase Acid Protein (Pa) From Influenza A Virus, Wilson-Smith/1933 (H1N1) Bound To Follow On Fragment Ebsi-4723 4-(5-Chlorothiophen-2-Yl)-1H-Pyrazole
Organism: Influenza a virus (strain a/wilson-smith/1933 h1n1)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-03-08 Classification: TRANSCRIPTION Ligands: 8QG |
 |
Crystal Structure Of Carveol Dehydrogenase From Mycobacterium Avium In Complex With Nad
Organism: Mycobacterium avium
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-12-02 Classification: OXIDOREDUCTASE Ligands: NAD, PO4, MPD |
 |
Crystal Structure Of 2-C-Methyl-D-Erythritol 4-Phosphate Cytidylyltransferase (Ispd) From Burkholderia Thailandensis Complexed With Ctp
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-04-29 Classification: TRANSFERASE Ligands: CTP, MG, ACT, CAD, GOL |
 |
Crystal Structure Of 2-C-Methyl-D-Erythritol 4-Phosphate Cytidylyltransferase (Ispd) From Burkholderia Thailandensis
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2015-04-01 Classification: TRANSFERASE Ligands: GOL, ACT |
 |
Crystal Structure Of A Putative Carveol Dehydrogenase From Mycobacterium Avium Bound To Nad
Organism: Mycobacterium avium
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-10-15 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of Putative 2-Dehydropantoate 2-Reductase Pane From Mycobacterium Tuberculosis Complexed With Nadp And Oxamate
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-02-26 Classification: OXIDOREDUCTASE Ligands: NAP, OXM, ACT, CL, GOL |

