Search Count: 22
 |
Crystal Structure Of Saccharomyces Cerevisiae Nup192, Residues 2 To 960 [Scnup192(2-960)]
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2013-02-20 Classification: PROTEIN TRANSPORT Ligands: SO4, IOD |
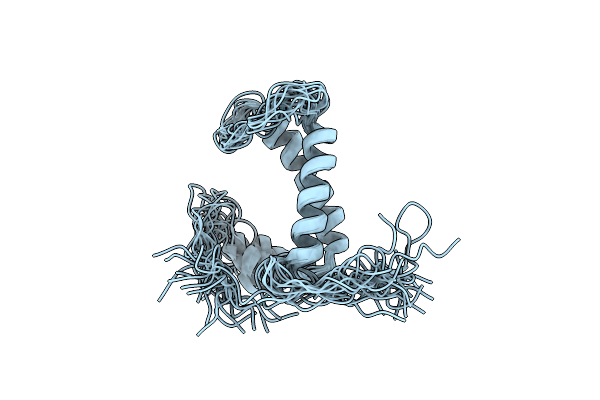 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2012-05-23 Classification: MEMBRANE PROTEIN |
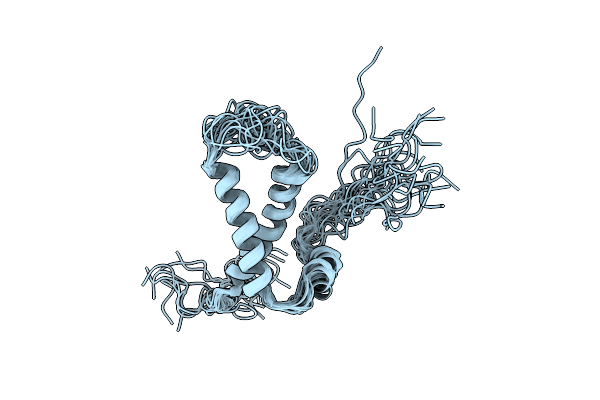 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2012-05-23 Classification: MEMBRANE PROTEIN |
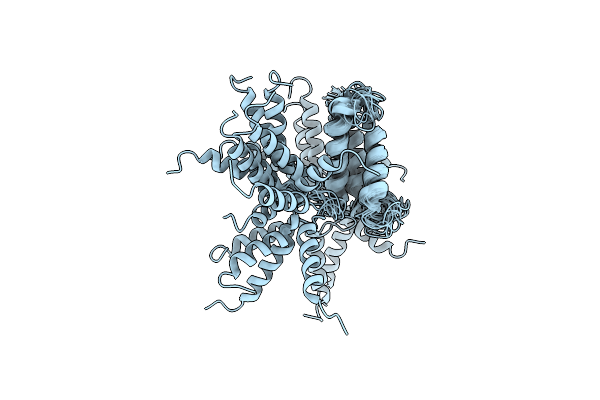 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2012-05-23 Classification: MEMBRANE PROTEIN |
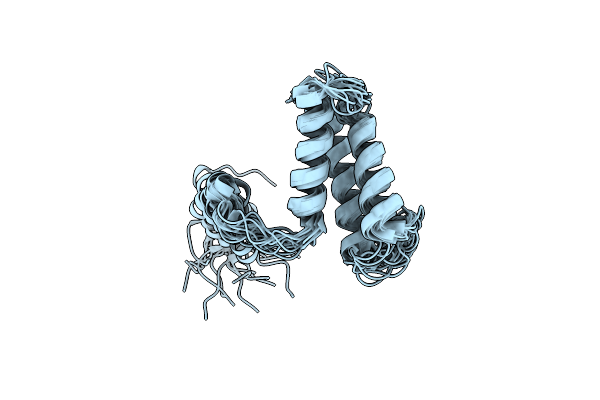 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2012-05-23 Classification: MEMBRANE PROTEIN |
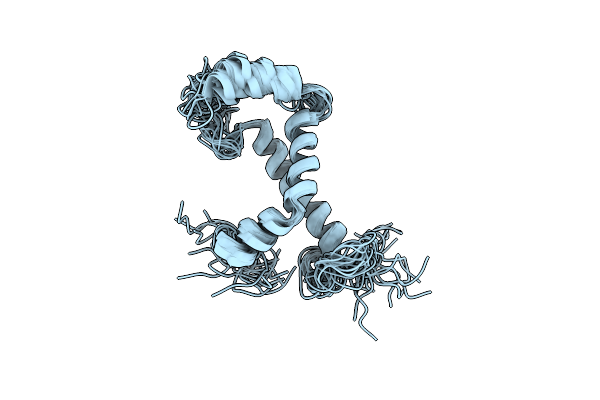 |
Backbone Structure Of Human Membrane Protein Fam14B (Interferon Alpha-Inducible Protein 27-Like Protein 1)
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2012-05-23 Classification: MEMBRANE PROTEIN |
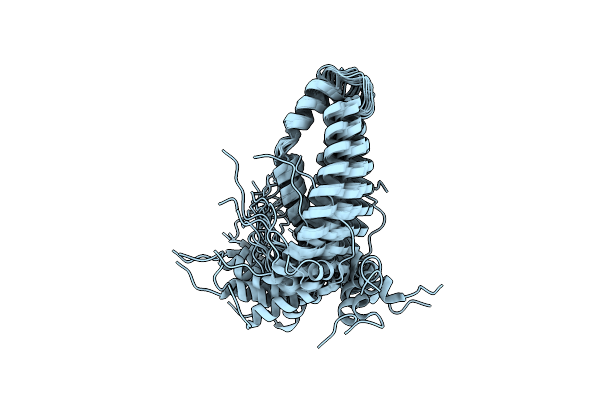 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2012-05-23 Classification: MEMBRANE PROTEIN |
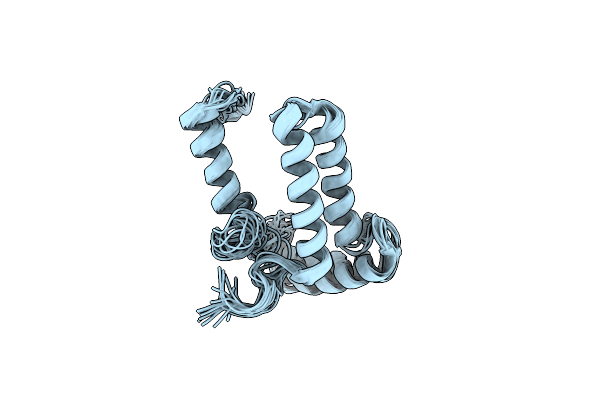 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2012-05-23 Classification: MEMBRANE PROTEIN |
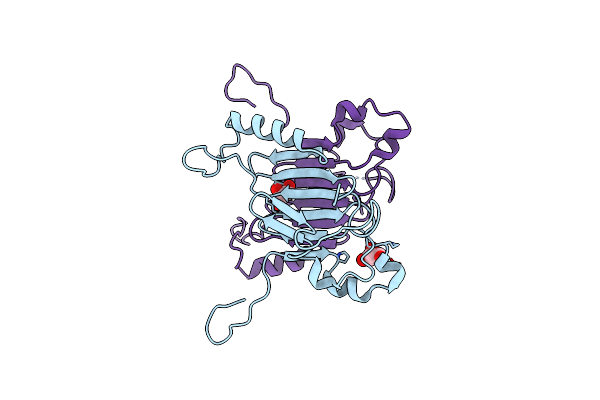 |
Crystal Structure Of The C-Terminal Domain Of Nuclear Pore Complex Component Nup116 From Candida Glabrata
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2010-08-04 Classification: PROTEIN TRANSPORT Ligands: GOL |
 |
Crystal Structure Of The C-Terminal Domain From The Nuclear Pore Complex Component Nup133 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-01-26 Classification: PROTEIN TRANSPORT Ligands: GOL |
 |
Crystal Structure Of The Autoproteolytic Domain From The Nuclear Pore Complex Component Nup145 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2009-12-22 Classification: PROTEIN TRANSPORT, RNA BINDING PROTEIN Ligands: EDO |
 |
Crystal Structure Of The Autoproteolytic Domain From The Nuclear Pore Complex Component Nup145 From Saccharomyces Cerevisiae In The Hexagonal, P61 Space Group
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-12-22 Classification: PROTEIN TRANSPORT, RNA BINDING PROTEIN Ligands: EDO |
 |
Organism: Pseudomonas fluorescens pf-5
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2009-10-20 Classification: TRANSPORT PROTEIN Ligands: LDA |
 |
Crystal Structure Of Mandelate Racemase/Muconate Lactonizing Enzyme From Bacillus Subtilis Complexed With Mg++ At 1.8 A
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2006-04-18 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: MG, CL |
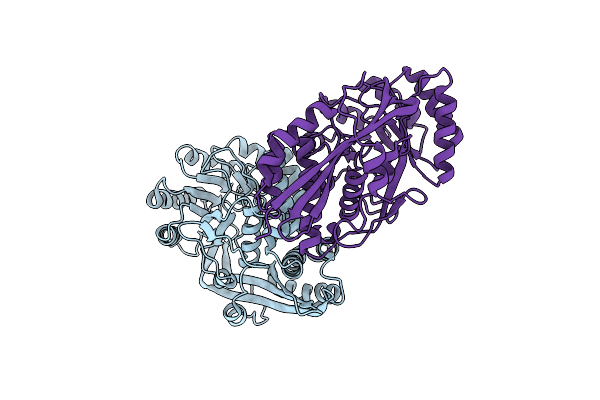 |
Crystal Structure Of Mandelate Racemase/Muconate Lactonizing Enzyme From Bacillus Subtilis At 1.8 A Resolution
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-04-04 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-08-22 Classification: OXIDOREDUCTASE, LYASE Ligands: FE2 |
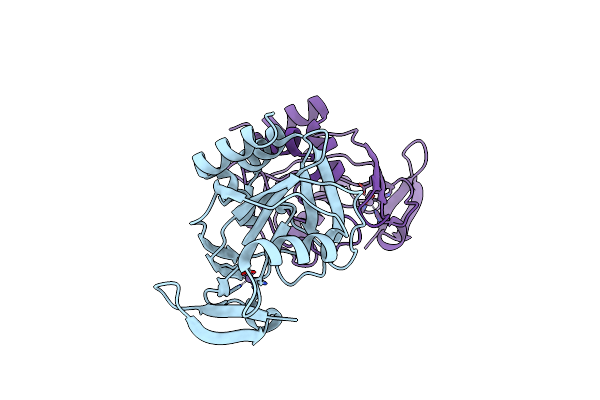 |
Structural Studies Of Cholesterol Biosynthesis: Mevalonate 5-Diphosphate Decarboxylase And Isopentenyl Diphosphate Isomerase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2001-03-28 Classification: ISOMERASE Ligands: MN |
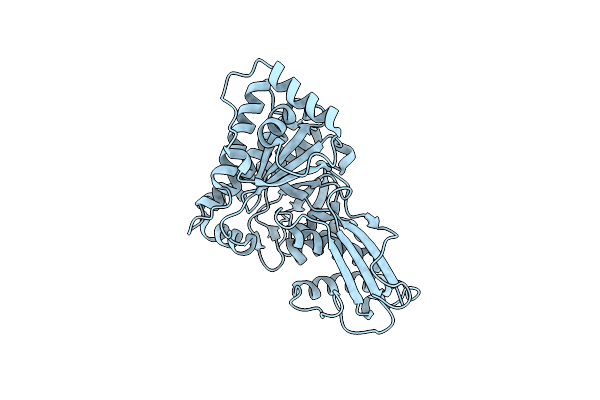 |
The X-Ray Crystal Structure Of Mevalonate 5-Diphosphate Decarboxylase At 2.3 Angstrom Resolution.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2001-03-21 Classification: LYASE |
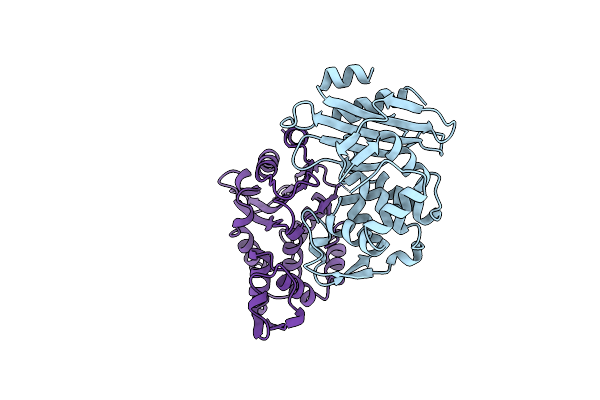 |
Crystal Structure Of The Omega Loop Deletion Mutant (Residues 163-178 Deleted) Of Beta-Lactamase From Staphylococcus Aureus Pc1
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1998-05-27 Classification: HYDROLASE Ligands: CL |
 |
Organism: Haloferax volcanii
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 1998-02-25 Classification: OXIDOREDUCTASE Ligands: PO4 |

