Search Count: 61
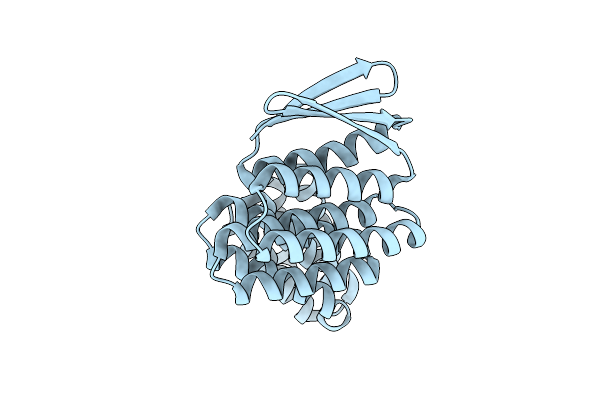 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: CL |
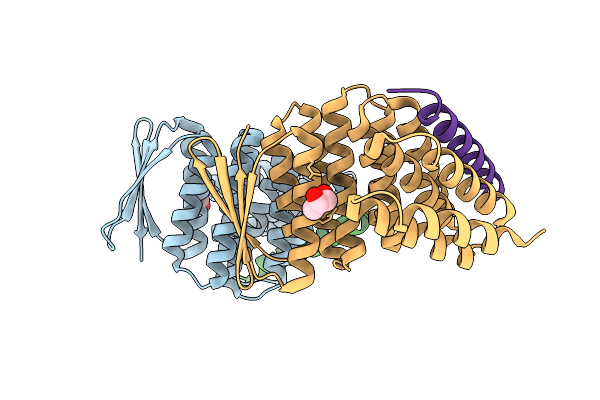 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As1 In Complex State He
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: PGO |
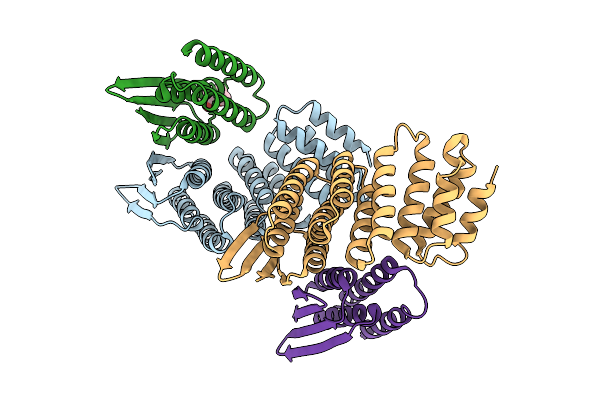 |
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State Th With Methylated Lysines, Crystal #1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: MPD |
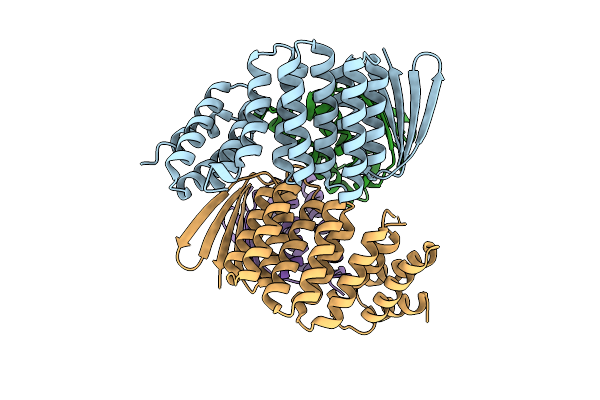 |
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State Th With Methylated Lysines, Crystal #2
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.88 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State The With Methylated Lysines
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
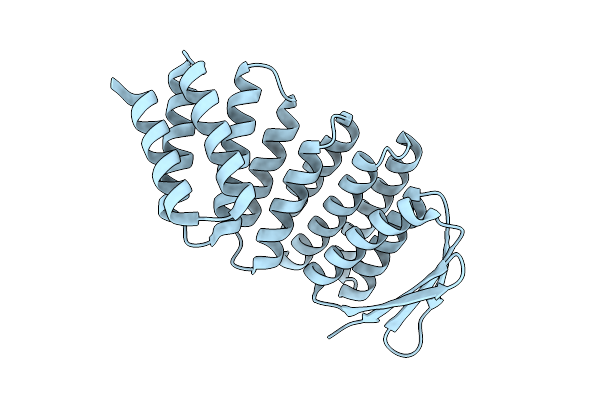 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As5
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
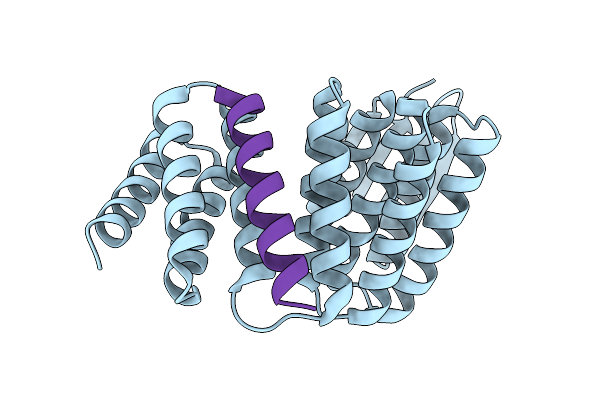 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As5 In Complex State He
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
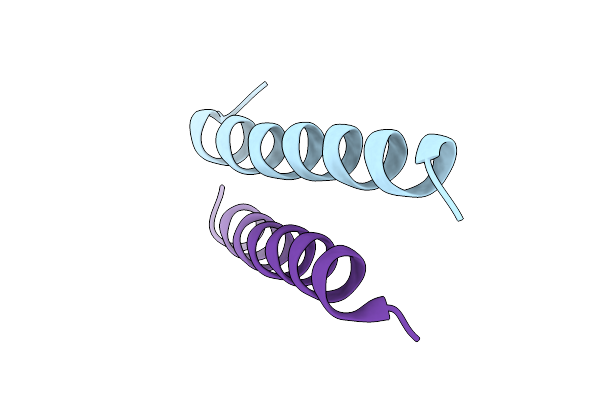
Crystal Structure Of Designed Facilitated Dissociation Target Lhd101An1: Lhd101A With An N-Terminal Extension
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
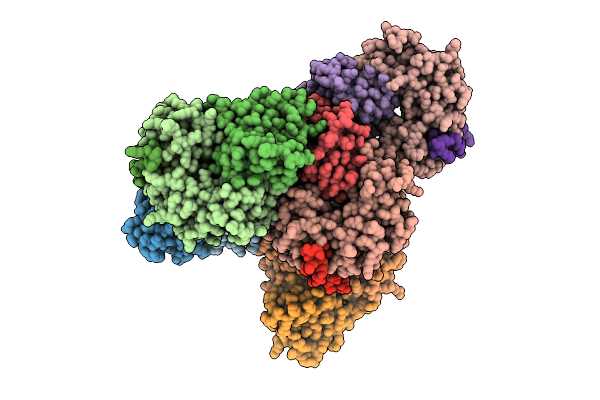
Crystal Structure Of Designed Conformational Switch Effector Peptide Cs221B
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Designed Allosteric Facilitated Dissociation Switch As1_K46L_E50W_K172W_E173Y In Complex State The
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.48 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
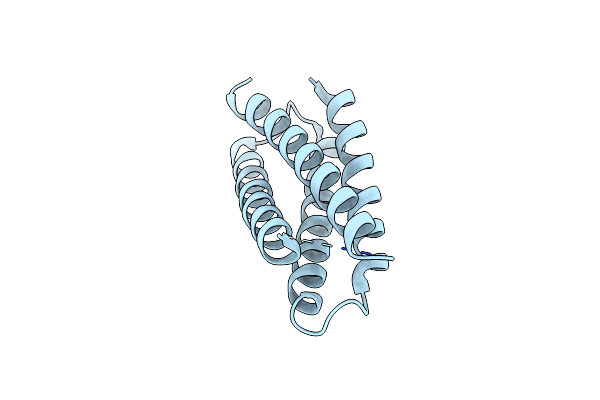
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: IMMUNE SYSTEM |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-10-23 Classification: DE NOVO PROTEIN |
 |
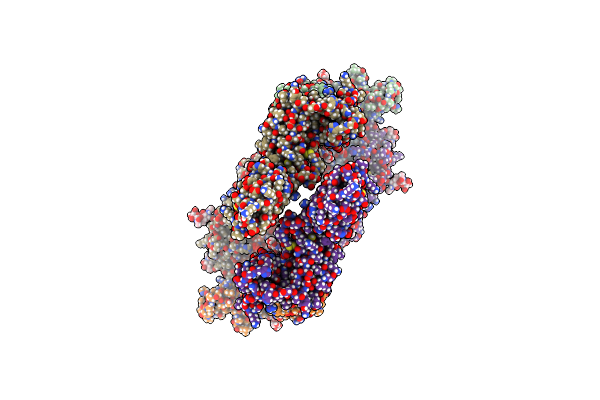
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: SIGNALING PROTEIN Ligands: NAG |
 |
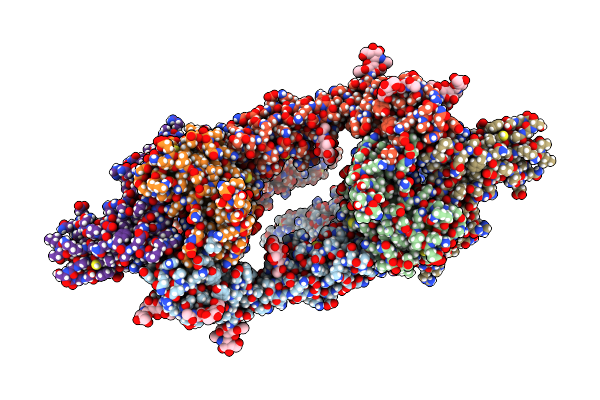
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
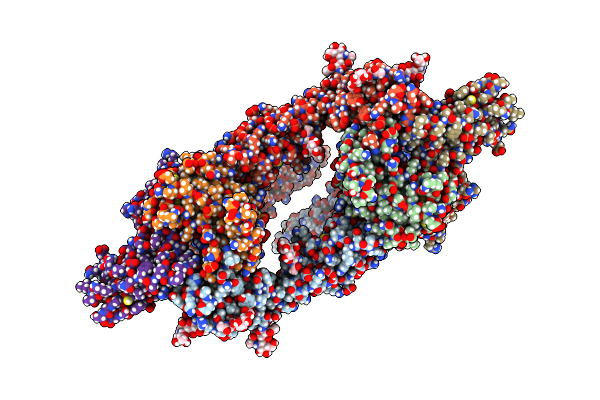
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: CYTOKINE Ligands: NAG, MAN |
 |
Crystal Structure For The Fniii Module Of Mouse Lep-R In Complex With The Anti-Lep-R Nanobody Vhh-4.80
Organism: Mus musculus, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-04-05 Classification: CYTOKINE Ligands: NAG, SO4 |
 |
Cryo-Em Structure For Mouse Leptin In Complex With The Mouse Lep-R Ectodomain (1:2 Mlep:Mlepr Model).
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: CYTOKINE |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: CYTOKINE Ligands: NI |

