Search Count: 12
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-03-05 Classification: HYDROLASE Ligands: CL, NA, GOL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-03-05 Classification: HYDROLASE Ligands: GOL, CL |
 |
Crystal Structure Of Susa Amylase From Bacteroides Thetaiotaomicron Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, IMD, CA |
 |
Crystal Structure Of Susg From Bacteroides Thetaiotaomicron Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, A1ILG, CA, IMD, ACT |
 |
Crystal Structure Of Amylase 5 (Amy5) From Ruminococcus Bromii Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Ruminococcus bromii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, A1IHI, CA |
 |
Crystal Structure Of K38 Amylase From Bacillus Sp. Strain Ksm-K38 Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacillus sp. ksm-k38
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, ACT, NA |
 |
Crystal Structure Of Gh31 Family Sulfoquinovosidase Bmsqase In Covalent Complex With Sq-Aziridine (Sqz)
Organism: Priestia megaterium dsm 319
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: Y2W, K |
 |
Crystal Structure Of Nad-Dependent Glycoside Hydrolase From Flavobacterium Sp. (Strain K172) In Complex With Co-Factor Nad+ And Sulfoquinovose (Sq)
Organism: Paenarthrobacter ureafaciens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: NAD, R7R, TRS |
 |
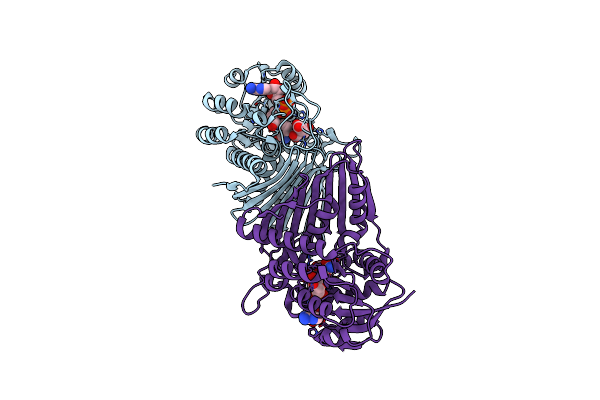
Crystal Structure Of Oxidoreductive Sulfoquinovosidase From Arthrobacter Sp. U41 (Arsqga)In Complex With Co-Factor Nad+
Organism: Arthrobacter sp. u41
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: NAD |
 |
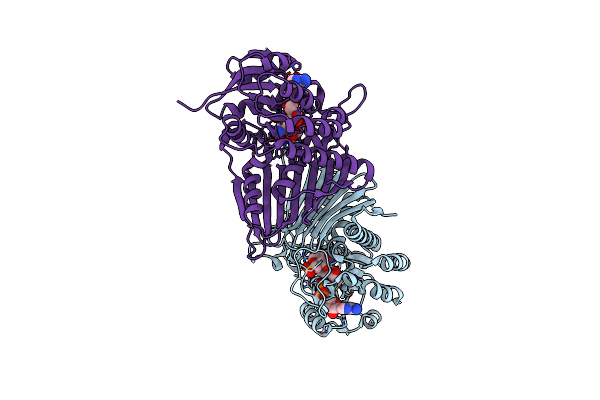
Crystal Structure Of Nad-Dependent Glycoside Hydrolase From Arthrobacter Sp. U41 In Complex With Nad+ Cofactor And Citrate
Organism: Arthrobacter sp. u41
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: NAD, CIT |
 |
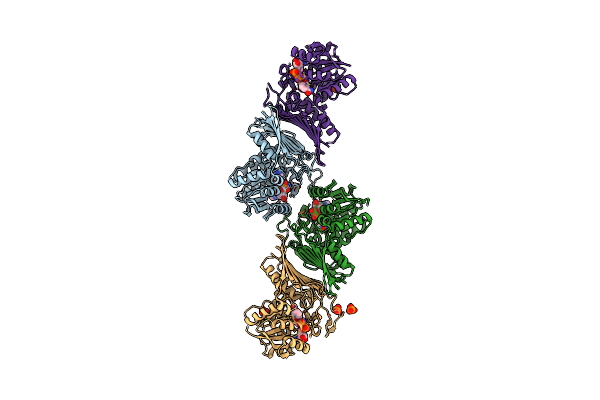
Crystal Structure Of Nad-Dependent Glycoside Hydrolase From Arthrobacter Sp. U41 In Complex With Nad+ And Sulfoquinovose (Sq)
Organism: Arthrobacter sp. u41
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: NAD, R7R |
 |
Crystal Structure Of Nad-Dependent Glycoside Hydrolase From Flavobacterium Sp. (Strain K172) In Complex With Co-Factor Nad+
Organism: Paenarthrobacter ureafaciens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: PO4, NAD |

