Search Count: 50
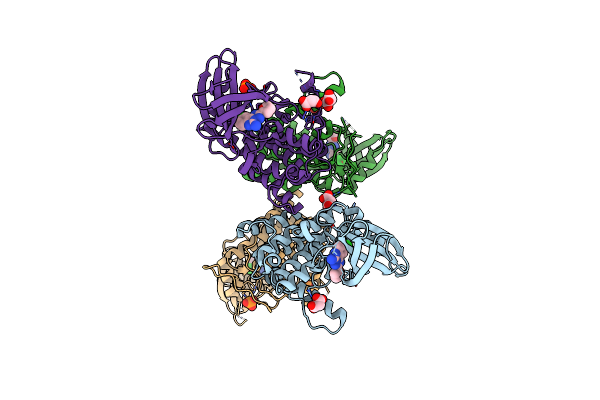 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: UX2, MLI, PO4 |
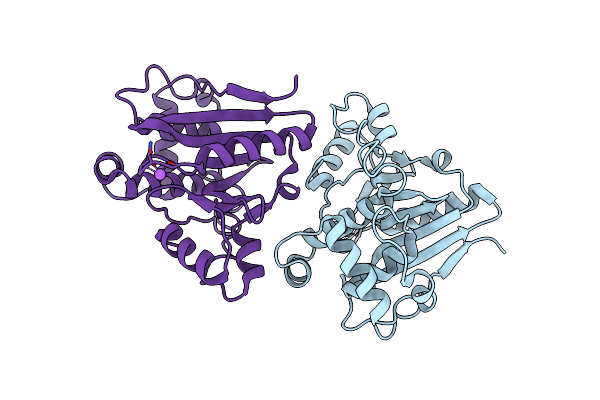 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-02-02 Classification: HYDROLASE |
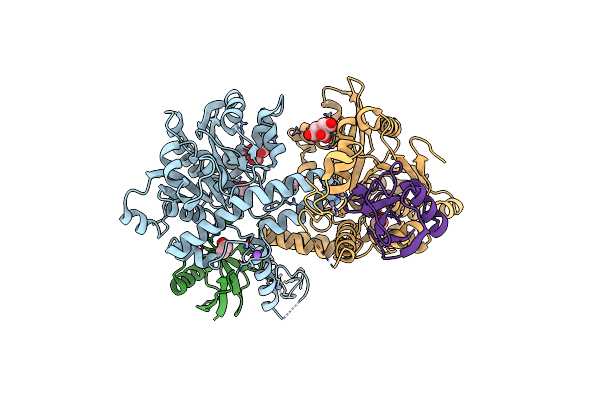 |
Crystal Structure Of The Covalent Complex Between Tribolium Castaneum Deubiquitinase Zup And Ubiquitin-Pa
Organism: Tribolium castaneum, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-02-02 Classification: HYDROLASE Ligands: CIT, EDO, ZN, NA |
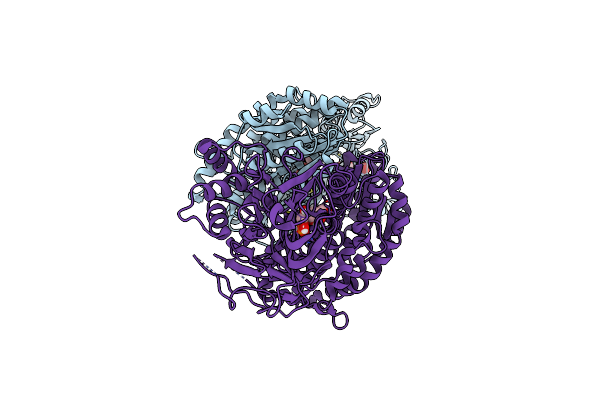 |
Crystal Structure Of Human Gfat-1 In Complex With Glucose-6-Phosphate And L-Glu
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: G6Q, GLU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: G6Q |
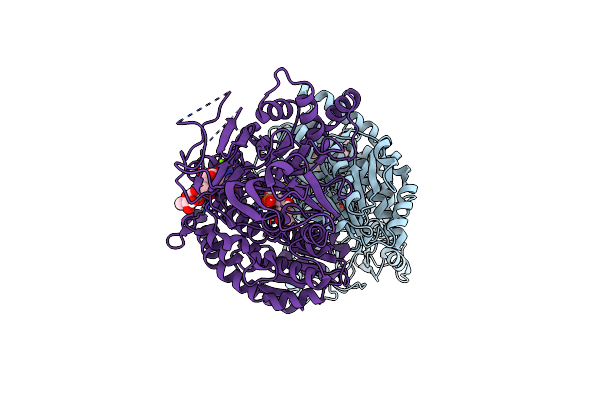 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: UD1, MG, G6Q |
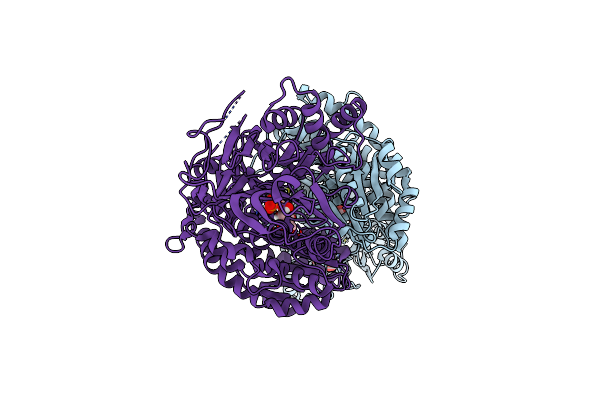 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: G6Q, GLU |
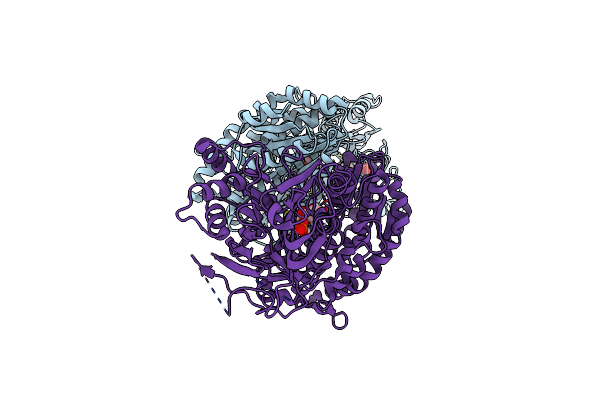 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: G6Q, GLU |
 |
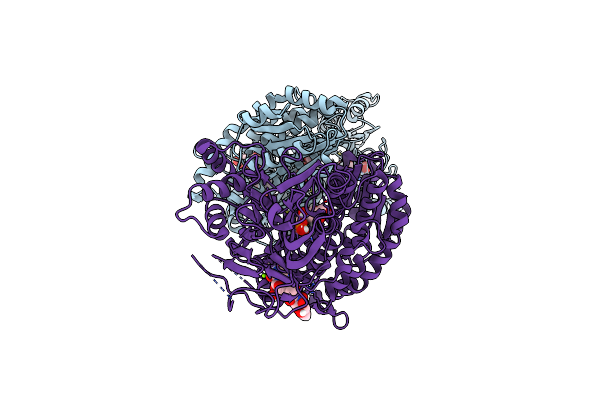
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: UD1, MG, G6Q, GLU |
 |
Crystal Structure Of Human Gfat-1 In Complex With Glucose-6-Phosphate, L-Glu, And Udp-Galnac
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: G6Q, GLU, UD2, MG |
 |
Crystal Structure Of Human Gfat-1 In Complex With Glucosamine-6-Phosphate And L-Glu
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: AGP, GLU |
 |
Crystal Structure Of Human Gfat-1 In Complex With Glucose-6-Phosphate, L-Glu, And Udp-Glcnac
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: UD1, MG, G6Q, GLU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: G6Q, GLU |
 |
Crystal Structure Of Human Casein Kinase I Delta In Complex With A Photoswitchable 2-Azoimidazole-Based Inhibitor (Compound 3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2019-09-11 Classification: TRANSFERASE Ligands: GEW, PO4 |
 |
Crystal Structure Of Human Casein Kinase I Delta In Complex With A Photoswitchable 2-Azothiazole-Based Inhibitor (Compound 2)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2019-09-11 Classification: TRANSFERASE Ligands: GE5, MLA |
 |
3D Nmr Solution Structure Of Ligand Peptide (Ac)Evnppvp Of Pro-Pro Endopeptidase-1
Organism: Clostridioides difficile
Method: SOLUTION NMR Release Date: 2019-06-19 Classification: HYDROLASE |
 |
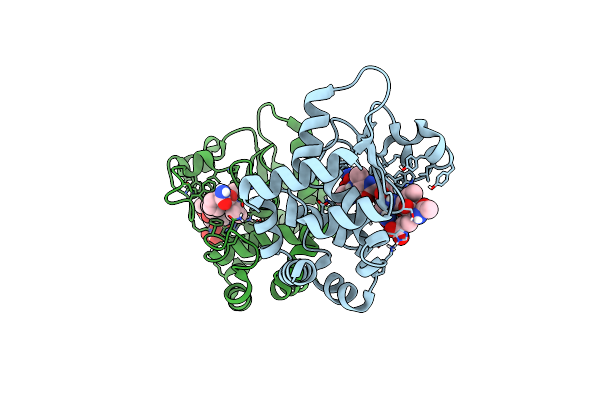
Crystal Structure Of Apo Ppep-1(E143A/Y178F) In Complex With Substrate Peptide Ac-Evnapvp-Conh2
Organism: Peptoclostridium difficile, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN |
 |
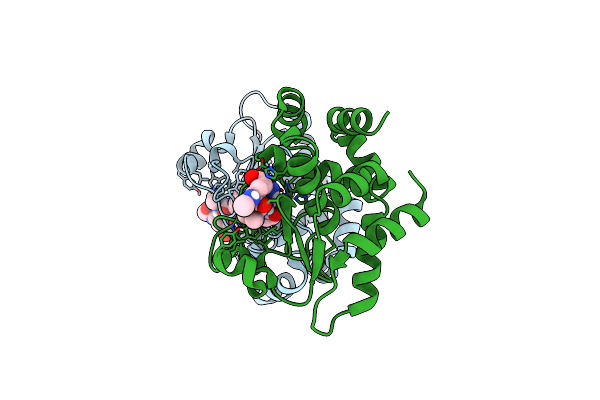
Crystal Structure Of Ppep-1(E143A/Y178F) In Complex With Substrate Peptide Ac-Evnpavp-Conh2
Organism: Peptoclostridium difficile, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN |
 |
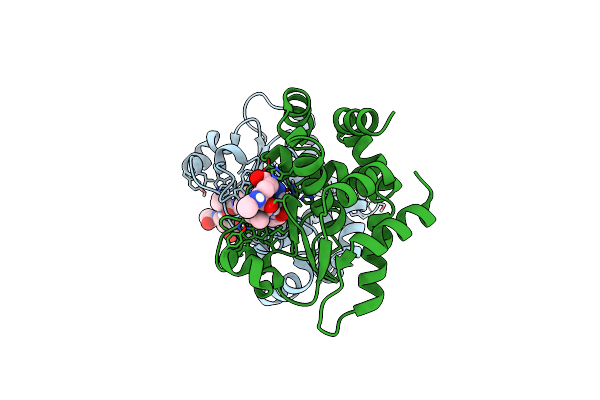
Crystal Structure Of Holo Ppep-1(E143A/Y178F) In Complex With Product Peptide Ac-Evnp-Co2 (Substrate Peptide: Ac-Evnpavp-Conh2)
Organism: Peptoclostridium difficile, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Holo Ppep-1(E143A/Y178F) In Complex With Product Peptide Ac-Evnp-Co2 (Substrate Peptide: Ac-Evnppvp-Conh2)
Organism: Peptoclostridium difficile, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN, NI |

