Search Count: 9
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: VIRAL PROTEIN Ligands: NAG |
 |
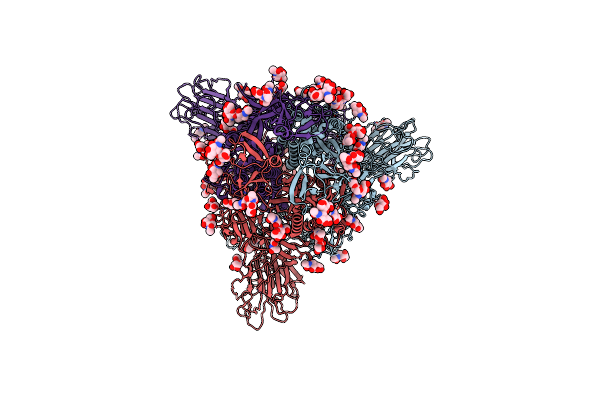
Cryo-Em Structure Of Sars-Cov-2 Spike In Complex With Neutralizing Antibody B1-182.1 That Targets The Receptor-Binding Domain
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-07-28 Classification: IMMUNE SYSTEM/VIRAL PROTEIN |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike In Complex With Neutralizing Antibody B1-182.1 That Targets The Receptor-Binding Domain
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-07-28 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike In Complex With Neutralizing Antibody A23-58.1 That Targets The Receptor-Binding Domain
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-07-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike In Complex With Neutralizing Antibody A23-58.1 That Targets The Receptor-Binding Domain
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-07-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
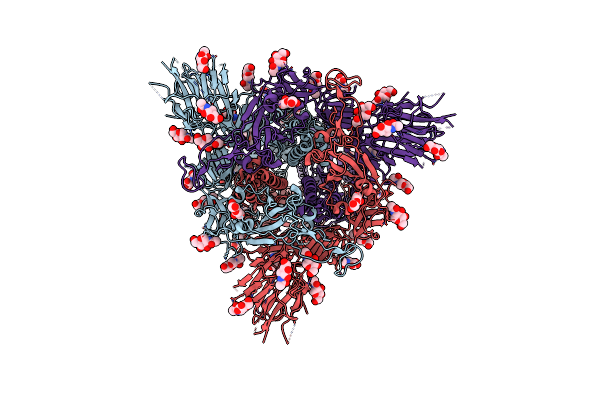 |
Cryo-Em Structure Of A Biotinylated Sars-Cov-2 Spike Probe In The Prefusion State (Rbds Down)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-09-02 Classification: VIRAL PROTEIN Ligands: NAG |
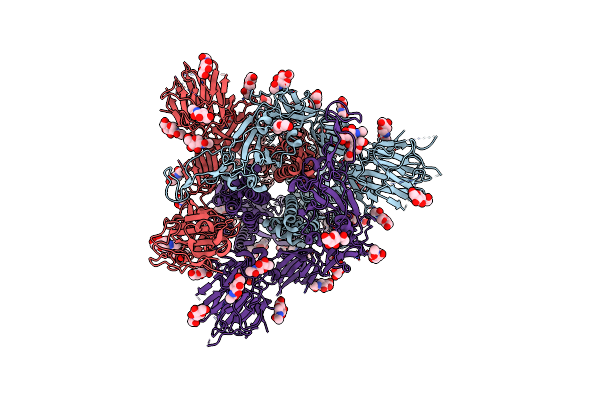 |
Cryo-Em Structure Of A Biotinylated Sars-Cov-2 Spike Probe In The Prefusion State (1 Rbd Up)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-09-02 Classification: VIRAL PROTEIN Ligands: NAG |
 |
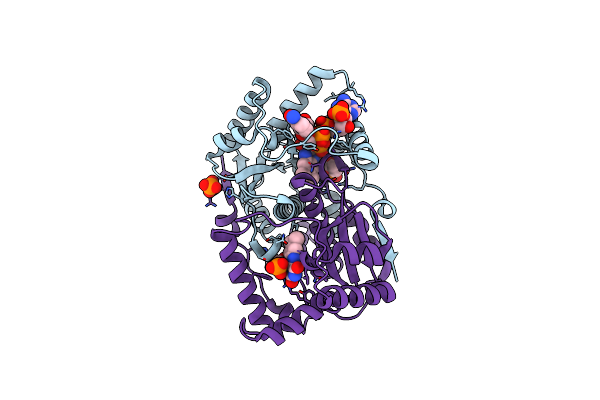
Crystal Structure Of The Fmn-Dependent Nitroreductase Nfnb From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2010-07-14 Classification: OXIDOREDUCTASE Ligands: FMN, PO4, GOL |
 |
Crystal Structure Of The Fmn-Dependent Nitroreductase Nfnb From Mycobacterium Smegmatis In Complex With Nadph
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-07-14 Classification: OXIDOREDUCTASE Ligands: FMN, NDP, PO4 |

