Search Count: 60
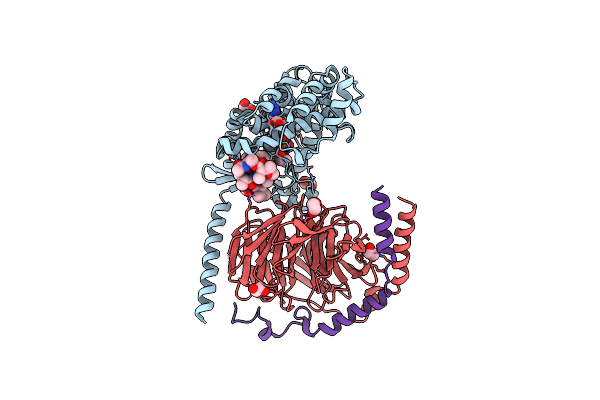 |
Crystal Structure Of The G11 Protein Heterotrimer Bound To Ym-254890 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-03-19 Classification: SIGNALING PROTEIN Ligands: GDP, EDO, DAM, HF2, THC, OTH, HL2, MAA, ALA, ACE |
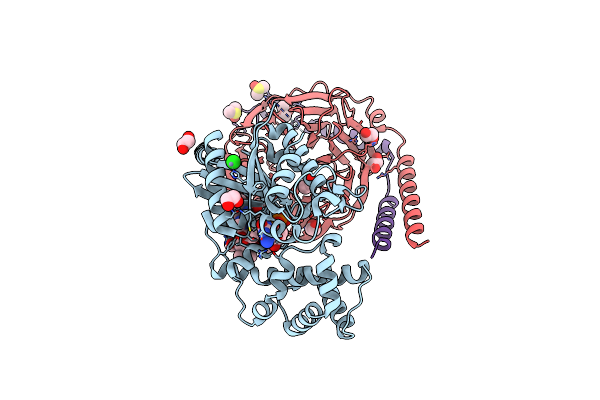 |
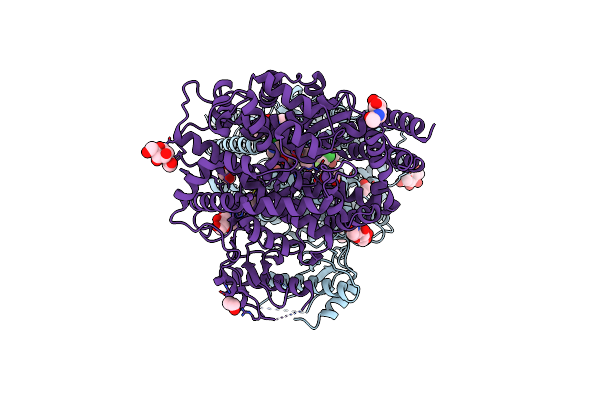
Crystal Structure Of The G11 Protein Heterotrimer Bound To Fr900359 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2025-03-19 Classification: SIGNALING PROTEIN Ligands: EDO, GDP, ZN, CL, DAM, HF2, UDL, OTH, HL2, MAA, ALA, PPI, DMS |
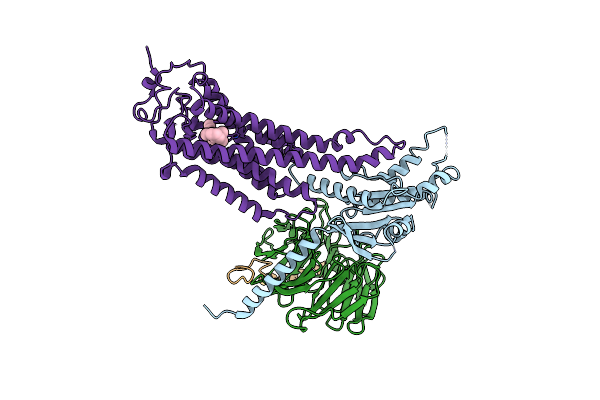 |
Organism: Homo sapiens, Hasarius adansoni, Bos taurus
Method: ELECTRON MICROSCOPY Resolution:4.90 Å Release Date: 2024-10-30 Classification: MEMBRANE PROTEIN Ligands: RET |
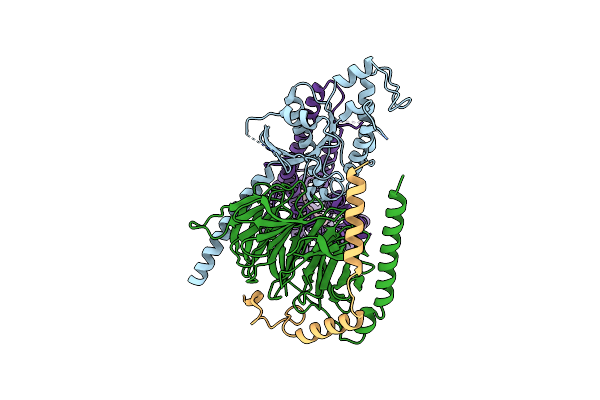 |
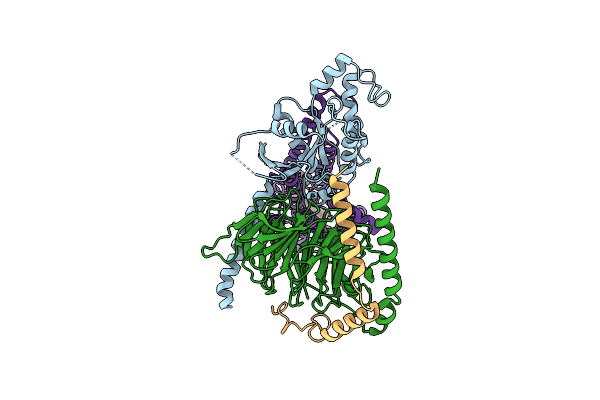
Cryo-Em Structure Of Jumping Spider Rhodopsin-1 Bound To A Giq Heterotrimer
Organism: Hasarius adansoni, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN Ligands: A1H6M |
 |
Cryo-Em Structure Of Jumping Spider Rhodopsin-1 Bound To A Giq Heterotrimer
Organism: Hasarius adansoni, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN Ligands: A1H6M |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: PROTEIN TRANSPORT Ligands: 9ED |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: PROTEIN TRANSPORT Ligands: 9ED |
 |
Structure Of Human Ace2 In Complex With A Fluorinated Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-07-17 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IDX, NAG, EDO, CL, NA, ZN |
 |
Intramembrane Recognition Between Transmembrane Domains Of Il-7R And Common Gamma Chain
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2023-07-05 Classification: MEMBRANE PROTEIN |
 |
Intramembrane Recognition Between Transmembrane Domains Of Il-9R And Common Gamma Chain
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2023-07-05 Classification: MEMBRANE PROTEIN |
 |
Dark State Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase (Sacla)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
 |
Dark State Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase (Swissfel)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
 |
1 Picosecond Light Activated Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
 |
10 Picosecond Light Activated Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
 |
100 Picosecond Light Activated Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase (Sacla)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: RET, NAG, DAO, OLC, PLM |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Mus musculus, Aliivibrio fischeri
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: IMMUNE SYSTEM |
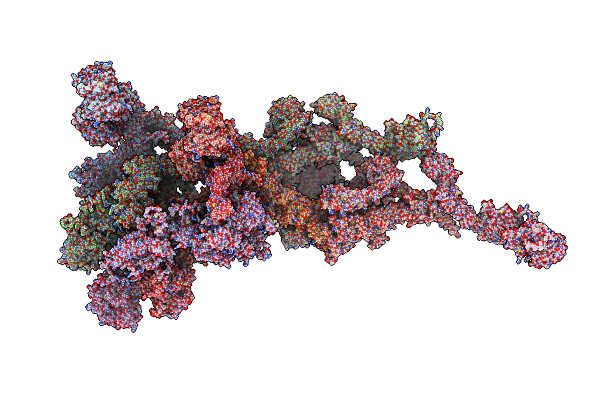 |
Cryo-Em Model Of Protomer Of The Cytoplasmic Ring Of The Nuclear Pore Complex From Xenopus Laevis
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: NUCLEAR PROTEIN |
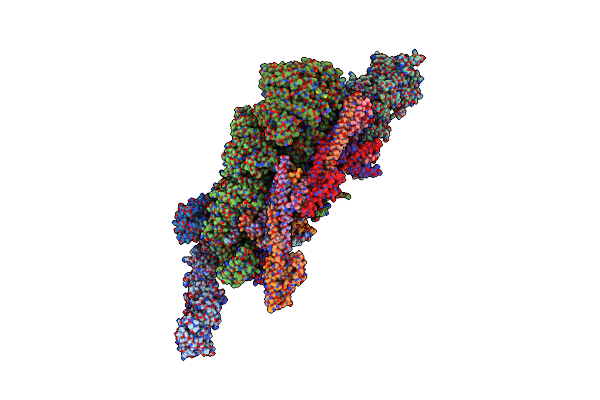 |
Cryo-Em Structure Of The Inner Ring Protomer Of The Saccharomyces Cerevisiae Nuclear Pore Complex
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-04-13 Classification: TRANSPORT PROTEIN |
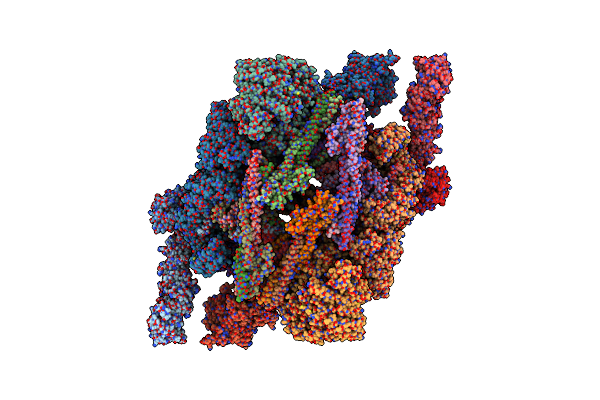 |
Cryo-Em Structure Of The Inner Ring Monomer Of The Saccharomyces Cerevisiae Nuclear Pore Complex
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-04-13 Classification: TRANSPORT PROTEIN |

