Search Count: 88
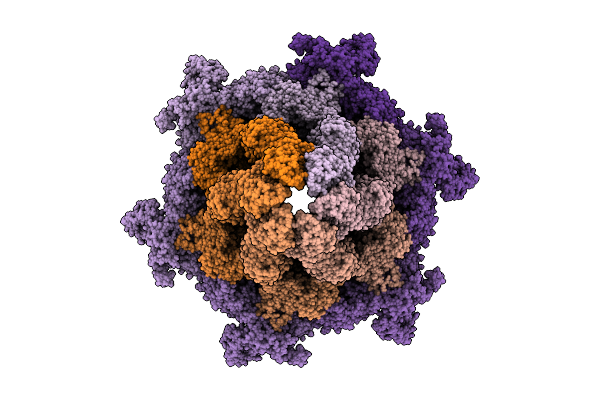 |
Cryo-Em Structure Of The Plpvc1 Baseplate, 6-Fold Symmetrized (C6), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
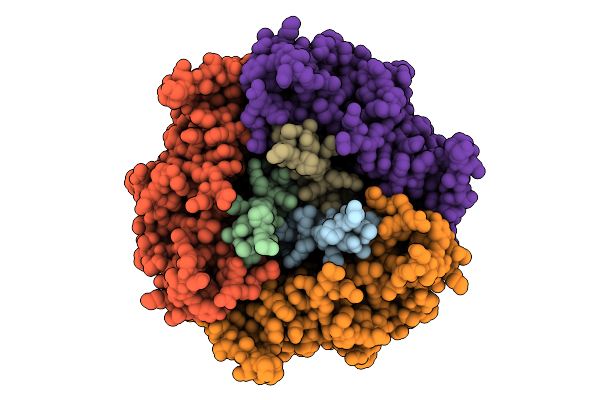 |
Cryo-Em Structure Of The Plpvc1 Central Spike, 3-Fold Symmetrized (C3), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
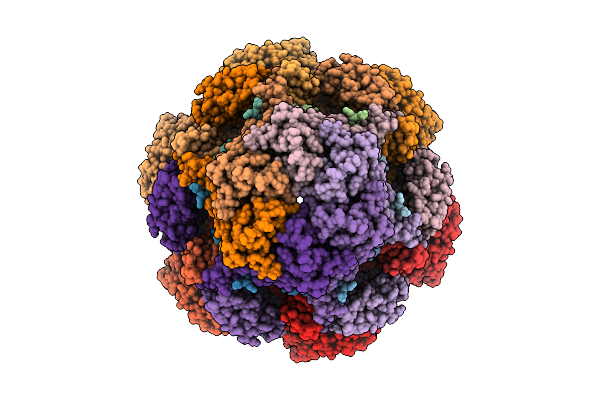 |
Cryo-Em Structure Of The Plpvc1 Cap, 6-Fold Symmetrized (C6), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
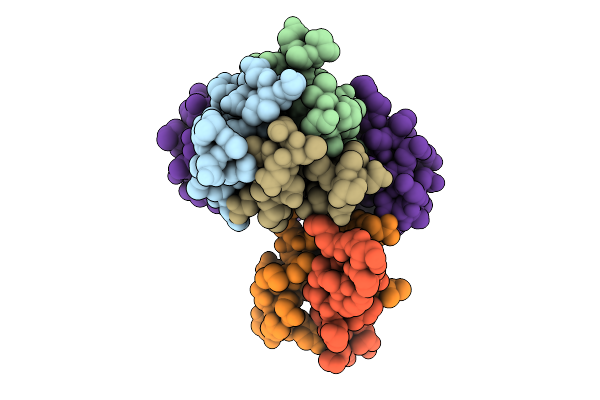 |
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
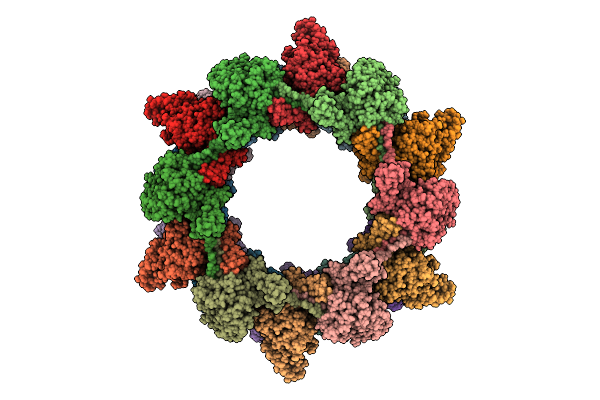 |
Cryo-Em Structure Of The Plpvc1 Sheath, 6-Fold Symmetrized (C6), In Contracted State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
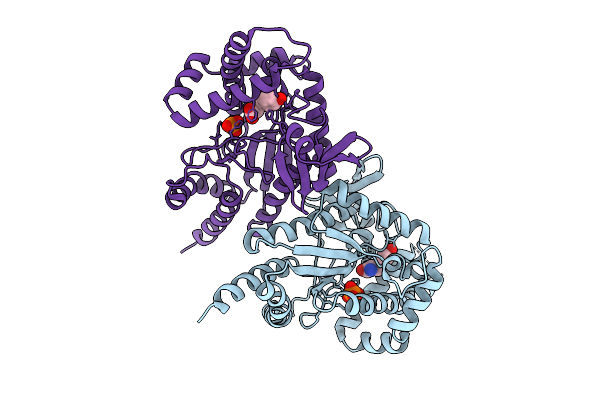 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PO4, TYR |
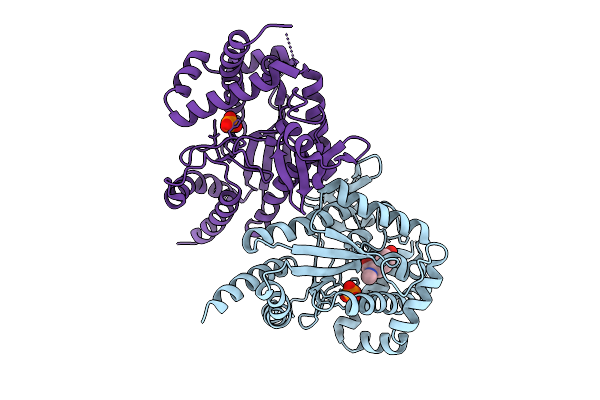 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PO4, I3J |
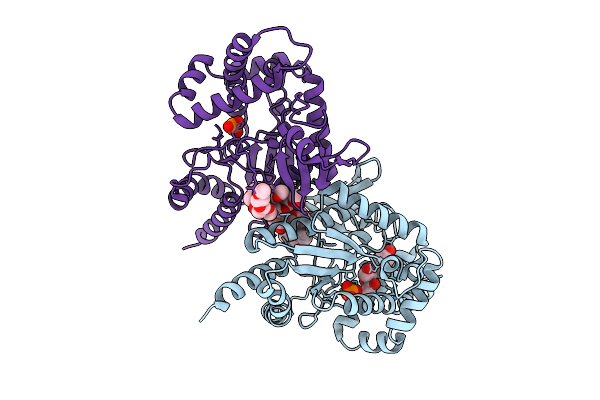 |
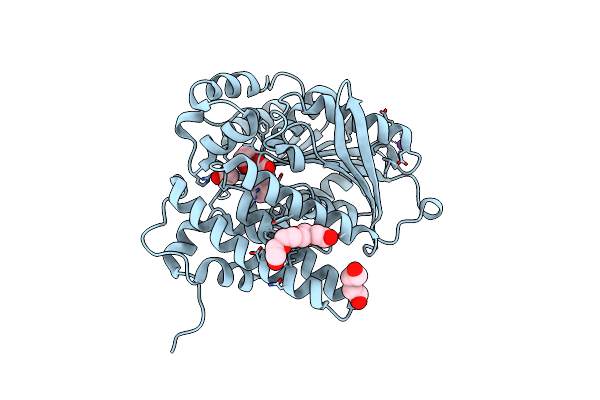
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PG4, A1A46, PO4, EDO, PG0, PG6 |
 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: LYASE Ligands: TCA, NA, EDO, PEG, PG4, ACT |
 |
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-13 Classification: TOXIN |
 |
Photorhabdus Luminescens Tcda1 Prepore-To-Pore Intermediate, K567W K2008W Mutant
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-13 Classification: TOXIN |
 |
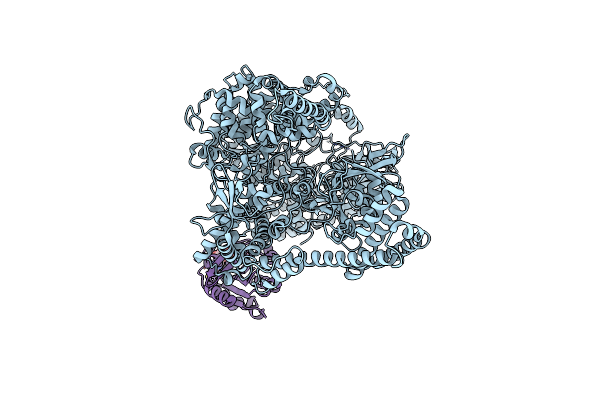
Photorhabdus Luminescens Tcda1 Prepore-To-Pore Intermediate, C16S, C20S, C870S, T1279C Mutant
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-13 Classification: TOXIN |
 |
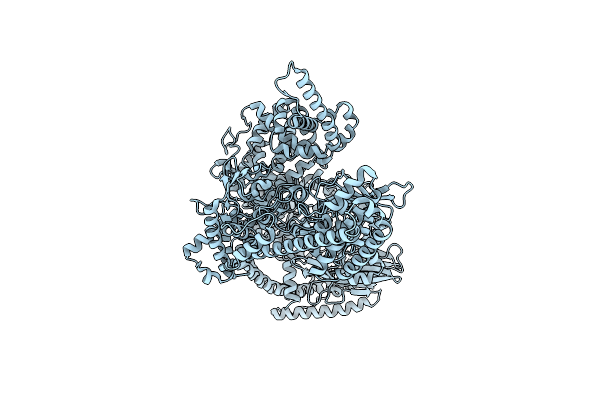
Photorhabdus Luminescens Makes Caterpillars Floppy (Mcf) Toxin With The C-Terminal Deletion In Complex With Arf3
Organism: Photorhabdus luminescens, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: TOXIN Ligands: GTP, MG |
 |
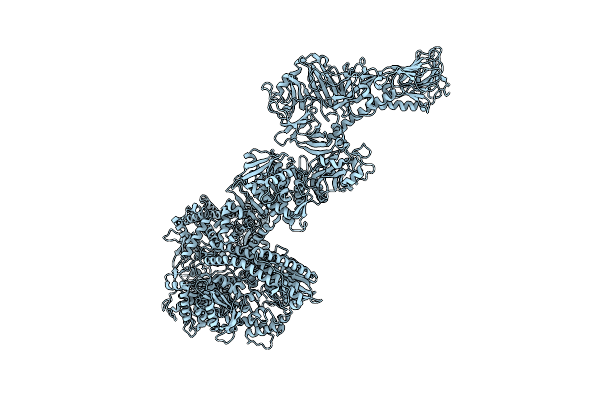
Photorhabdus Luminescens Makes Caterpillars Floppy (Mcf) Toxin With The C-Terminal Deletion
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: TOXIN |
 |
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: TOXIN |
 |
Organism: Photorhabdus luminescens, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: TOXIN Ligands: ADP, MG, APR, NCA |
 |
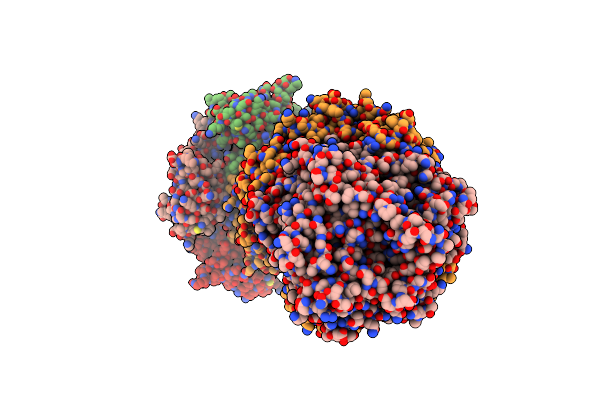
Structure Of The Adp-Ribosyltransferase Tccc3Hvr From Photorhabdus Luminescens
Organism: Photorhabdus luminescens
Method: SOLUTION NMR Release Date: 2022-06-29 Classification: TOXIN |
 |
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-31 Classification: TOXIN/PROTEIN TRANSPORT |
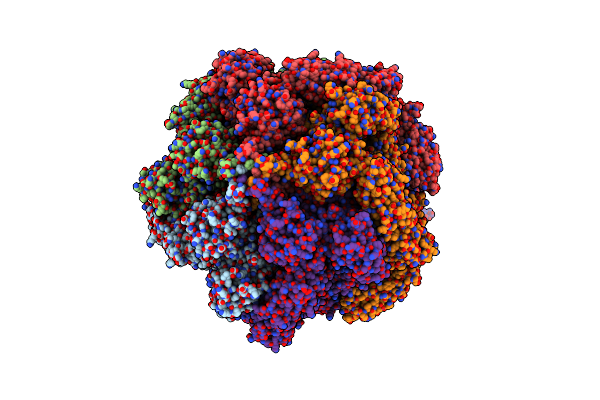 |
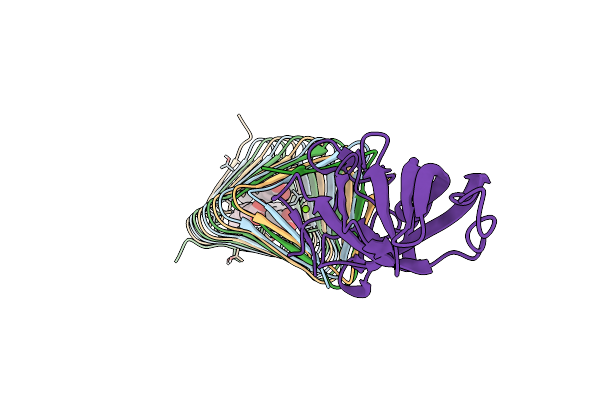
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2020-11-04 Classification: TOXIN |
 |
Photorhabdus Virulence Cassette (Pvc) Paar Repeat Protein Pvc10 In Complex With A T4 Gp5 Beta-Helix Fragment Modified To Mimic Pvc8, The Central Spike Protein Of Pvc
Organism: Enterobacteria phage t4, Photorhabdus luminescens subsp. laumondii (strain dsm 15139 / cip 105565 / tt01)
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: VIRAL PROTEIN Ligands: MG, STE, ELA, PLM |

