Search Count: 949
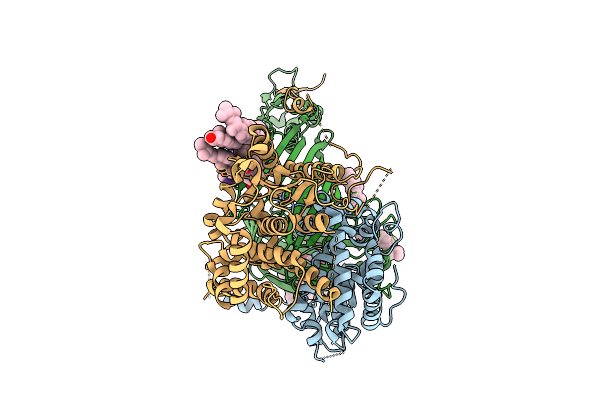 |
Organism: Thermothelomyces thermophilus, Photorhabdus khanii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN/ANTIBIOTIC Ligands: ERG |
 |
Apo Form Of Tumor Necrosis Factor-Like Lectin Pltl From Photorhabdus Laumondii
Organism: Photorhabdus laumondii subsp. laumondii tto1
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: SUGAR BINDING PROTEIN |
 |
Tumor Necrosis Factor-Like Lectin Pltl From Photorhabdus Laumondii In Complex With Blood Group B Trisaccharide
Organism: Photorhabdus laumondii subsp. laumondii tto1
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: SUGAR BINDING PROTEIN Ligands: FUC, GLA |
 |
Tumor Necrosis Factor-Like Lectin Pltl From Photorhabdus Laumondii In Complex With Lewis Y Tetrasaccharide
Organism: Photorhabdus laumondii subsp. laumondii tto1
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: SUGAR BINDING PROTEIN |
 |
Tumor Necrosis Factor-Like Lectin Pltl From Photorhabdus Laumondii In Complex With B Lewis B Pentasaccharide
Organism: Photorhabdus laumondii subsp. laumondii tto1
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: SUGAR BINDING PROTEIN |
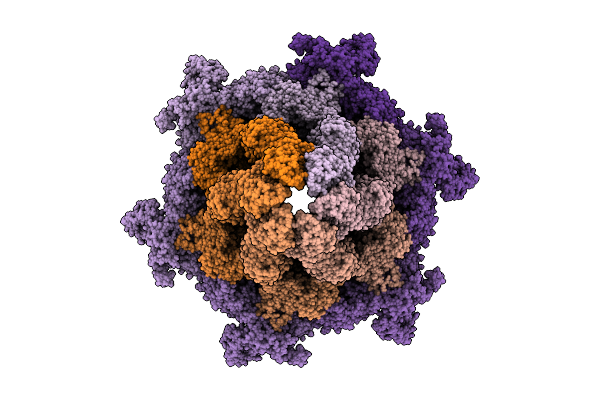 |
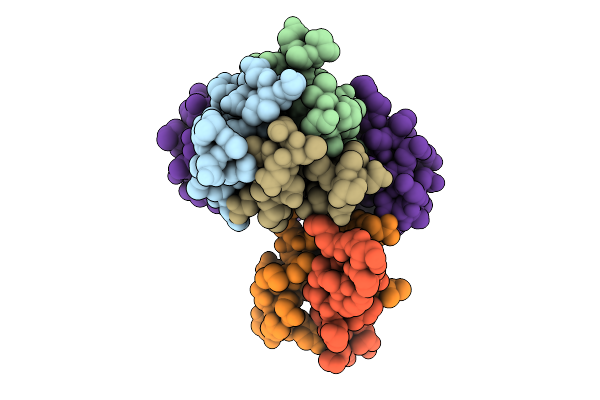
Cryo-Em Structure Of The Plpvc1 Baseplate, 6-Fold Symmetrized (C6), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
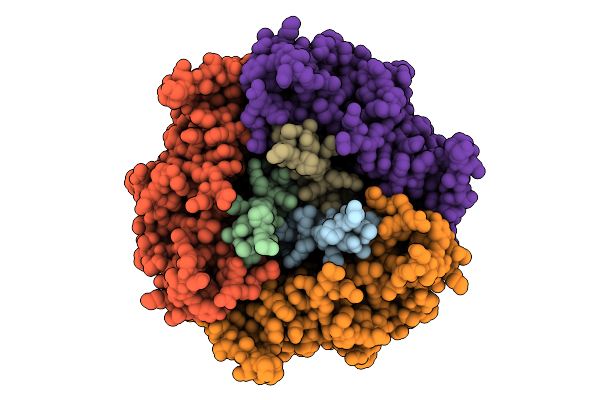 |
Cryo-Em Structure Of The Plpvc1 Central Spike, 3-Fold Symmetrized (C3), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
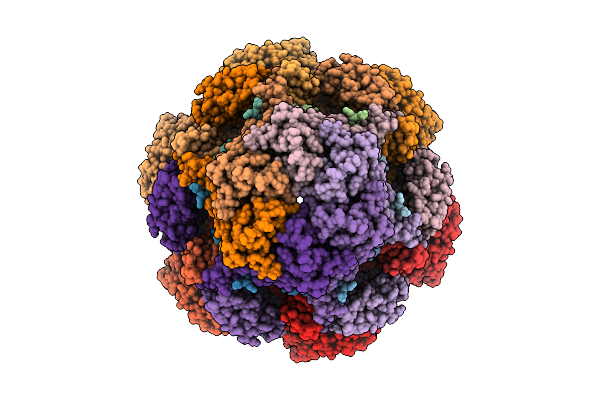 |
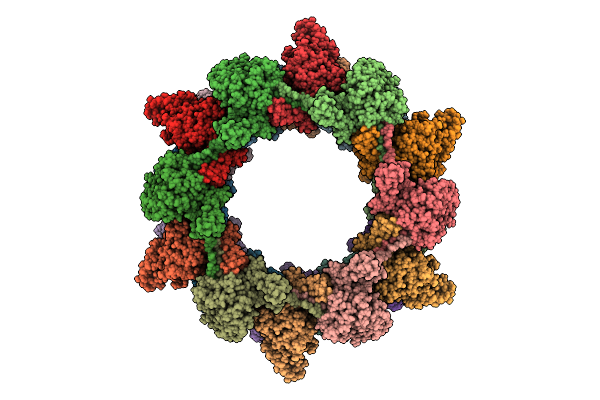
Cryo-Em Structure Of The Plpvc1 Cap, 6-Fold Symmetrized (C6), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of The Plpvc1 Sheath, 6-Fold Symmetrized (C6), In Contracted State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
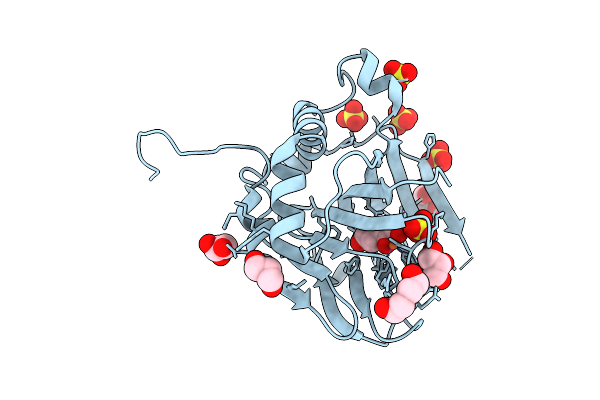 |
Organism: Photorhabdus asymbiotica
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: AKG, SO4, GOL, PEG, FE |
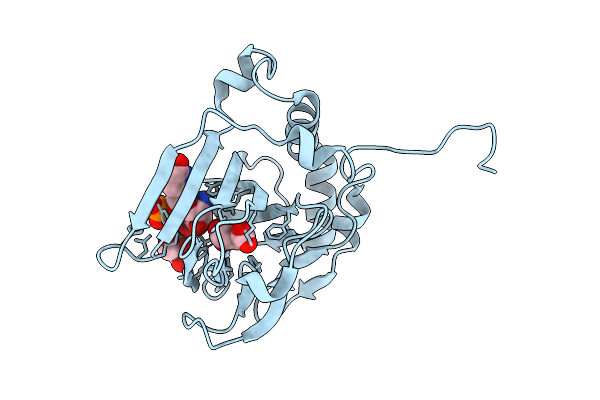 |
Pasi From Photorhabdus Asymbiotica Bound To Vanadyl, Succinate, And 5-Amino-6-Hydroxy-Octanosyl Acid 2-Phosphate
Organism: Photorhabdus asymbiotica
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: A1BIK, GOL, SIN, CL, V |
 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PO4, TYR |
 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PO4, I3J |
 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PG4, A1A46, PO4, EDO, PG0, PG6 |
 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: LYASE Ligands: TCA, NA, EDO, PEG, PG4, ACT |
 |
Organism: Escherichia coli, Synthetic construct, Photorhabdus khanii
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: 4NE |
 |
Organism: Escherichia coli, Synthetic construct, Photorhabdus khanii
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: 4NE |
 |
Organism: Escherichia coli k-12, Photorhabdus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: MG |
 |
Organism: Escherichia coli k-12, Photorhabdus
Method: ELECTRON MICROSCOPY Resolution:3.62 Å Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN |

