Search Count: 17
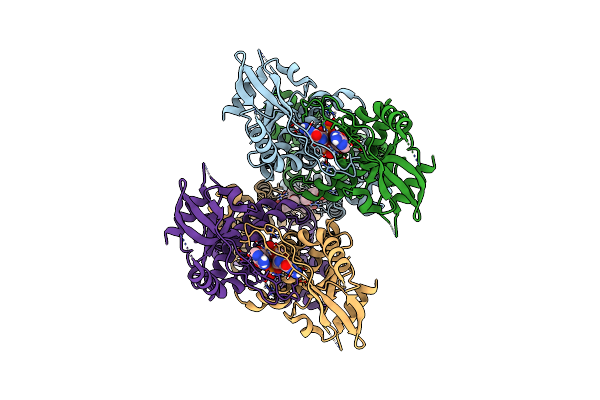 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: IMMUNE SYSTEM Ligands: 1SY, Y6H |
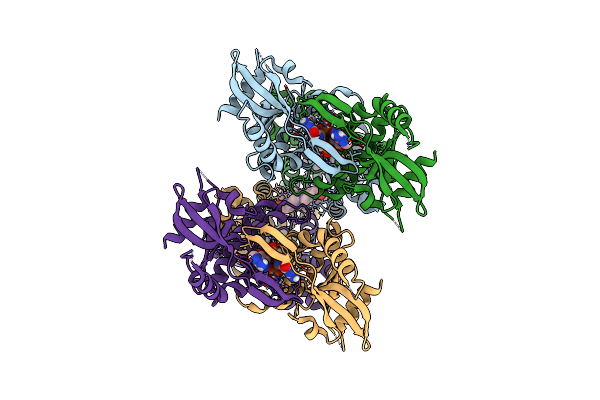 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: IMMUNE SYSTEM Ligands: Y6H, 1SY, 9IM |
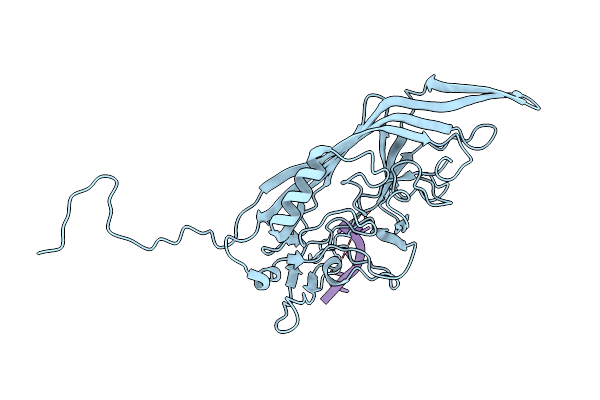 |
Organism: Unclassified densovirinae
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRUS/DNA |
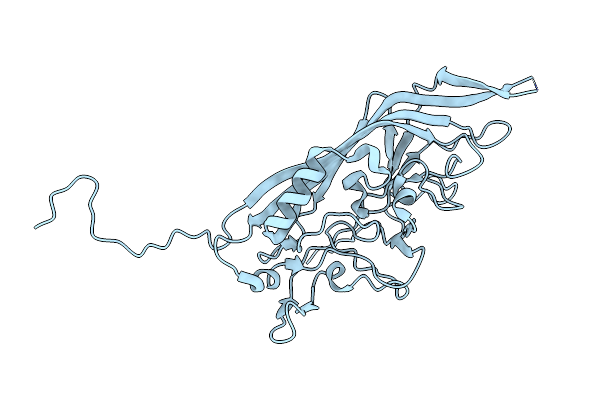 |
Acheta Domesticus Segmented Densovirus, High Buoyancy (Hb) Capsid, A Mixed Population Of Empty And Immature Full Particles
Organism: Unclassified densovirinae
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRUS |
 |
Acheta Domesticus Segmented Densovirus High Buoyancy Fraction 2 Empty Capsid Structure
Organism: Solenopsis invicta-associated densovirus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRUS LIKE PARTICLE |
 |
Acheta Domesticus Segmented Densovirus High Buoyancy Fraction 1 (Hb1) Empty Capsid Structure
Organism: Solenopsis invicta-associated densovirus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Solenopsis invicta-associated densovirus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Penaeus monodon metallodensovirus
Method: ELECTRON MICROSCOPY Release Date: 2020-04-22 Classification: VIRUS LIKE PARTICLE Ligands: CA |
 |
Capsid Structure Of Penaeus Monodon Metallodensovirus Following Edta Treatment
Organism: Penaeus monodon metallodensovirus
Method: ELECTRON MICROSCOPY Release Date: 2020-04-22 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.29 Å Release Date: 2019-06-19 Classification: TRANSCRIPTION ACTIVATOR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2019-06-19 Classification: TRANSCRIPTION ACTIVATOR |
 |
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2017-08-16 Classification: LIGASE Ligands: MN, BTN |
 |
Crystal Structure Of Lactococcus Lactis Pyruvate Carboxylase In Complex With Cyclic-Di-Amp
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-08-16 Classification: LIGASE Ligands: MN, ADP, MG, 2BA |
 |
Crystal Structure Of Lactococcus Lactis Pyruvate Carboxylase G746A Mutant In Complex With Cyclic-Di-Amp
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-08-16 Classification: LIGASE Ligands: MN, ADP, MG, 2BA |
 |
Crystal Structure Of Glucosyl-3-Phosphoglycerate Synthase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-06-06 Classification: TRANSFERASE Ligands: GOL |
 |
Crystal Structure Of Glucosyl-3-Phosphoglycerate Synthase From Mycobacterium Tuberculosis In Complex With Mg2+ And Uridine-Diphosphate (Udp)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-06-06 Classification: TRANSFERASE Ligands: UDP, GOL |
 |
Crystal Structure Of Glucosyl-3-Phosphoglycerate Synthase From Mycobacterium Tuberculosis In Complex With Mn2+, Uridine-Diphosphate (Udp) And Phosphoglyceric Acid (Pga)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2012-06-06 Classification: TRANSFERASE Ligands: UDP, MN, 3PG, PO4, GOL |

