Search Count: 69
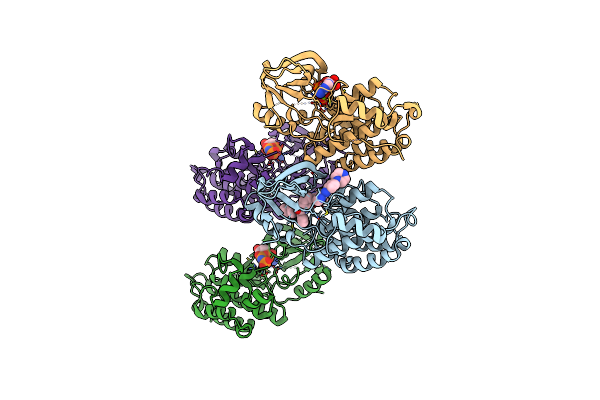 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: MG, ANP, IXC |
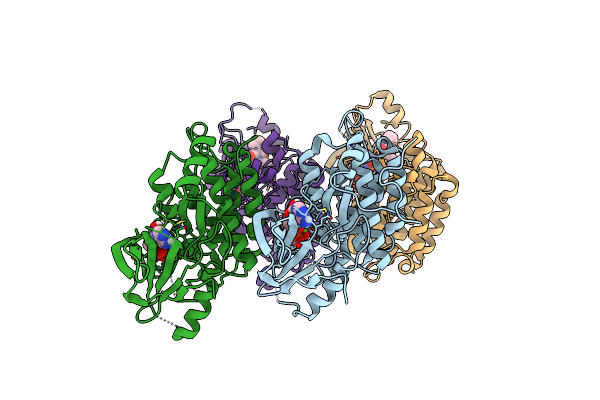 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: MG, ANP, IXR |
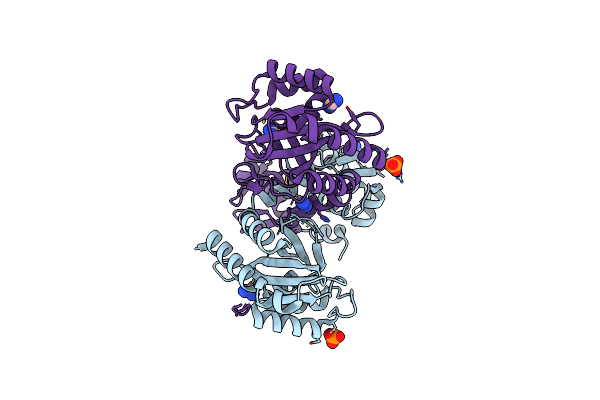 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: TRANSCRIPTION Ligands: SEY, PO4, CL |
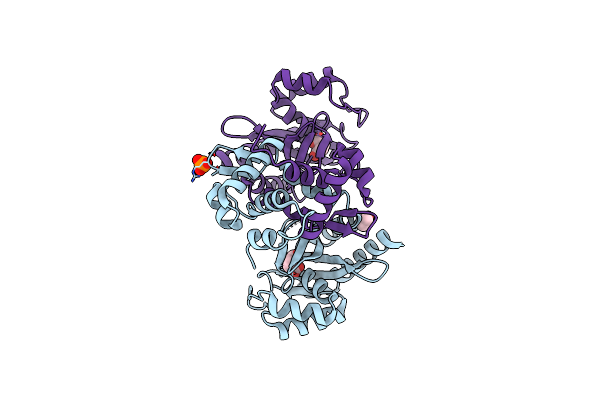 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: TRANSCRIPTION Ligands: SIN, EDO, PO4, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-17 Classification: TRANSFERASE Ligands: MG, ANP, YA6 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-01-17 Classification: TRANSFERASE Ligands: YAA, MG, ANP |
 |
Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Catalytically Active Radical State Solved By Xfel
Organism: Mesoplasma florum l1
Method: X-RAY DIFFRACTION Release Date: 2023-11-01 Classification: OXIDOREDUCTASE |
 |
Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Radical-Lost Ground State
Organism: Mesoplasma florum l1
Method: X-RAY DIFFRACTION Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: CA, GOL |
 |
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: FLC, PG4, PEG, PGE |
 |
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352) Bound To Neu5Ac2En (Dana)
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: DAN, PGE, PEG, PG4 |
 |
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352) Bound To Neu5Ac (Nana)
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: SIA, PGE, PEG, PG4 |
 |
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352)- Fructose Bound In Cbm
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: FLC, PEG, PGE, FUD |
 |
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352) D219A Mutant Bound To 3'-Sialyllactose (Only Neu5Ac Visible)
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: SIA, PEG, PGE, GAL |
 |
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352) D219A Mutant Bound To 6'-Sialyllactose (Only Neu5Ac Visible)
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: SIA, PEG, PGE, EDO, GAL |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul1 In Complex With 6-Hydroxymethylpterin
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR, GOL, SO4, CL |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: SO4, CL, GOL, PEG |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul2 In Complex With 7,8-Dihydropteroate, Magnesium, And Pyrophosphate
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: 78H, POP, MG, GOL, CL, PAB |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: GOL, CL, SO4 |
 |
Crystal Structure Of Sulfonamide Resistance Enzyme Sul3 In Complex With 6-Hydroxymethylpterin
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR |
 |
Crystal Structure Of Adaptive Laboratory Evolved Sulfonamide-Resistant Dihydropteroate Synthase (Dhps) From Escherichia Coli In Complex With 6-Hydroxymethylpterin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2023-05-03 Classification: TRANSFERASE Ligands: HHR |

