Search Count: 8
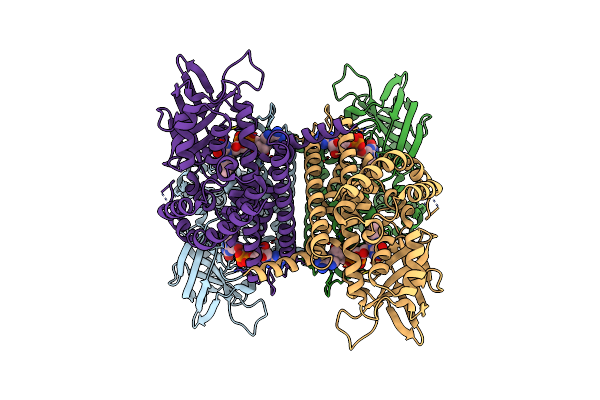 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: OXIDOREDUCTASE Ligands: FAD |
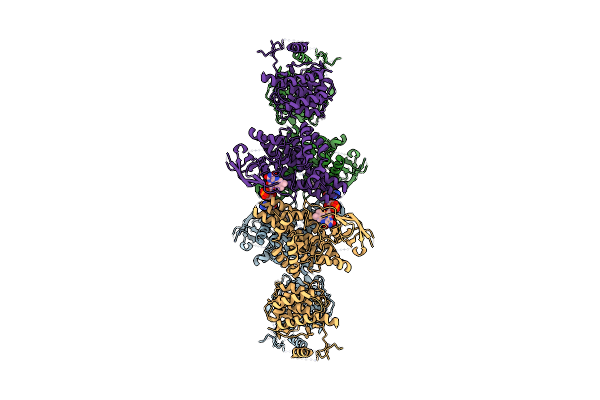 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-04-26 Classification: HYDROLASE Ligands: CL |
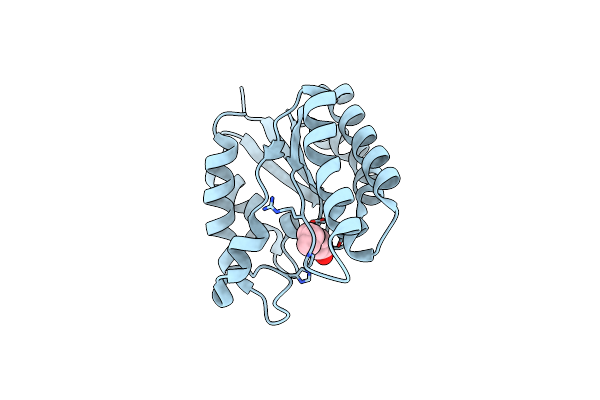 |
X-Ray Structure Of Acyl-Coa Thioesterase I, Tesa, Mutant M141L/Y145K/L146K At Ph 5 In Complex With Octanoic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2017-04-26 Classification: HYDROLASE Ligands: OCA |
 |
X-Ray Structure Of Acyl-Coa Thioesterase I, Tesa, Mutant M141L/Y145K/L146K At Ph 7.5 In Complex With Octanoic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2017-04-26 Classification: HYDROLASE Ligands: OCA |
 |
X-Ray Structure Of Acyl-Coa Thioesterase I, Tesa, Triple Mutant M141L/Y145K/L146K In Complex With Octanoic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2017-04-26 Classification: HYDROLASE Ligands: OCA, PEG |
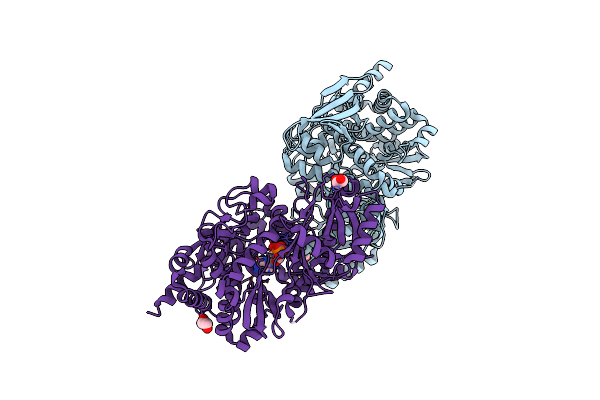 |
Crystal Structure Of Petrobactin Biosynthesis Protein Asbb From Bacillus Anthracis Str. Sterne
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2011-10-05 Classification: BIOSYNTHETIC PROTEIN Ligands: CL, EDO, ATP, MG |
 |
Crystal Structure Of The Probable 3-Dhs Dehydratase Asbf Involved In The Petrobactin Synthesis From Bacillus Anthracis
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2008-09-02 Classification: LYASE Ligands: MN, CL, DHB, TRS, GOL |

