Search Count: 16
 |
Symmetry-Expanded Reconstruction Of Augmin T-Ii Bonsai On The Gtpgammas Microtubule
Organism: Xenopus laevis, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CELL CYCLE Ligands: GTP, MG, GSP |
 |
Organism: Xenopus laevis, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: CELL CYCLE Ligands: G2P, MG, GTP |
 |
Organism: Xenopus laevis, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: CELL CYCLE Ligands: GTP, MG, G2P |
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: CELL CYCLE Ligands: G2P, MG, GTP |
 |
Organism: Xenopus laevis
Method: SOLID-STATE NMR Release Date: 2023-06-28 Classification: CELL CYCLE |
 |
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: CELL CYCLE |
 |
Crystal Structure Of Human Glucose Transporter Glut3 Bound With Exofacial Inhibitor Sa47
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-05-18 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: A0E, OLC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-05-18 Classification: TRANSPORT PROTEIN Ligands: OLC, GLC |
 |
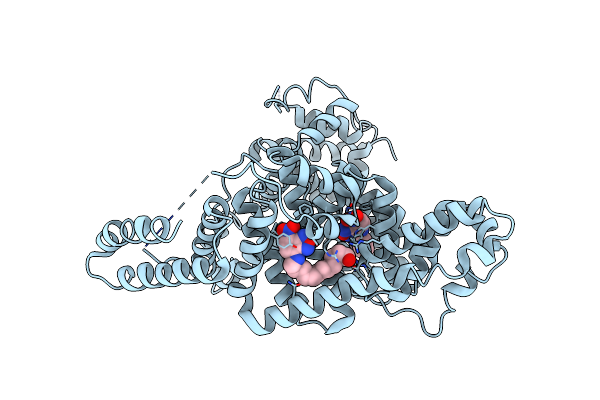
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2020-04-08 Classification: LIPID BINDING PROTEIN Ligands: DMS, SO4, SAS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2017-12-13 Classification: TRANSPORT PROTEIN Ligands: C7K |
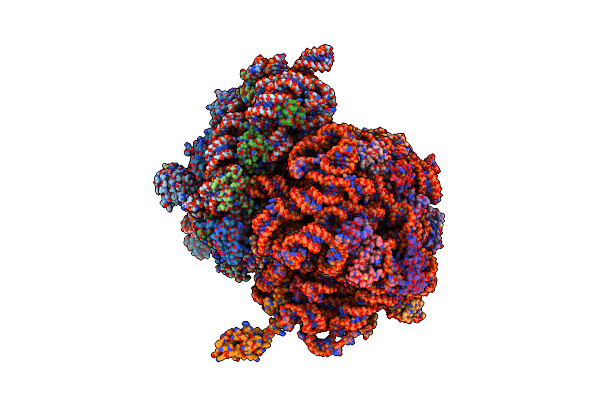 |
Organism: Escherichia coli, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:5.90 Å Release Date: 2014-07-09 Classification: RIBOSOME |
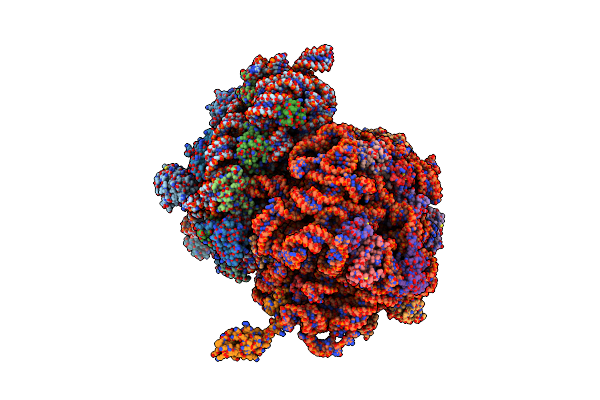 |
Organism: Escherichia coli, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:6.76 Å Release Date: 2014-07-09 Classification: RIBOSOME |
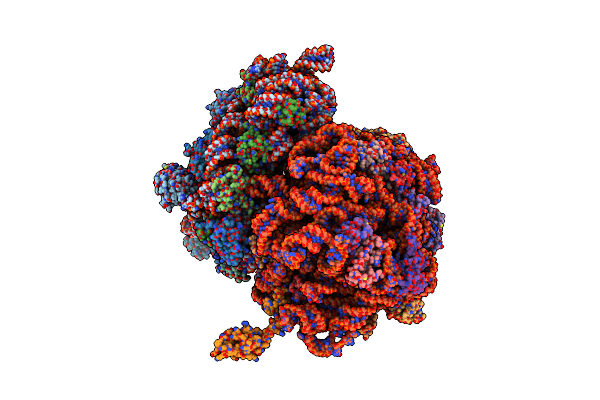 |
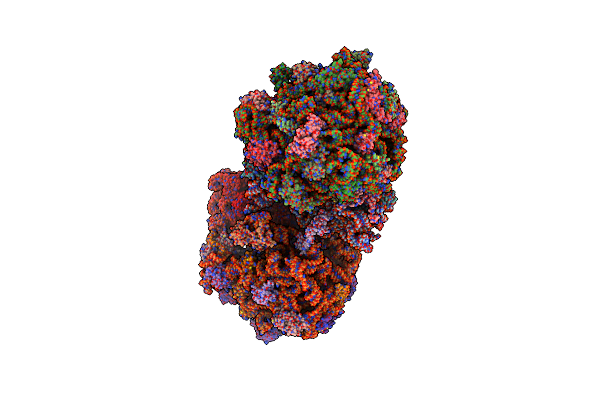
Crystal Structure Of The Whole Ribosomal Complex With A Stop Codon In The A-Site.
Organism: Escherichia coli, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:6.46 Å Release Date: 2014-07-09 Classification: RIBOSOME |
 |
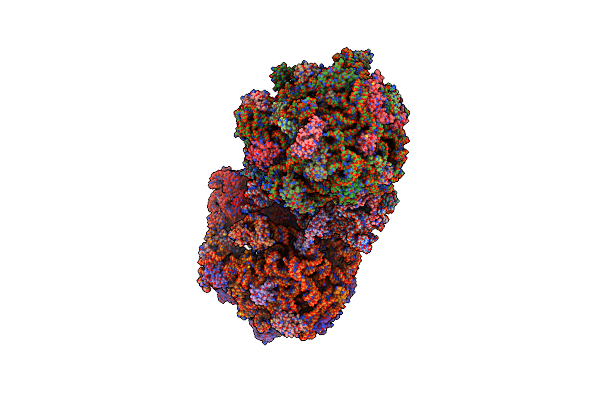
Structure Of The Thermus Thermophilus 70S Ribosome Complexed With Mrna, Trna And Paromomycin
Organism: Escherichia coli k-12 , Synthetic construct, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: MG, PAR, ZN |
 |
Structure Of The Ribosome Recycling Factor Bound To The Thermus Thermophilus 70S Ribosome With Mrna, Asl-Phe And Trna-Fmet
Organism: Escherichia coli, Synthetic construct, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: MG, ZN |
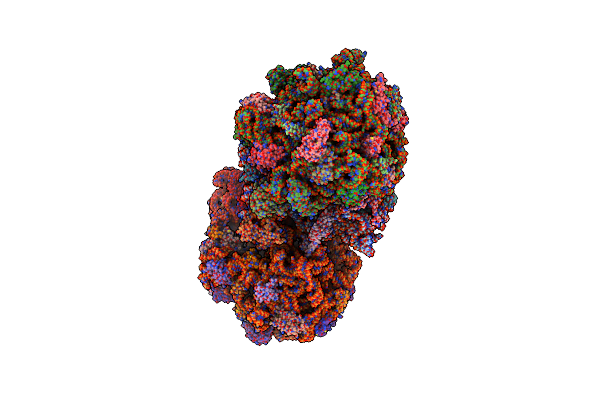 |
Insights Into Translational Termination From The Structure Of Rf2 Bound To The Ribosome
Organism: Escherichia coli k-12, Synthetic construct, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: MG, ZN |

