Search Count: 21
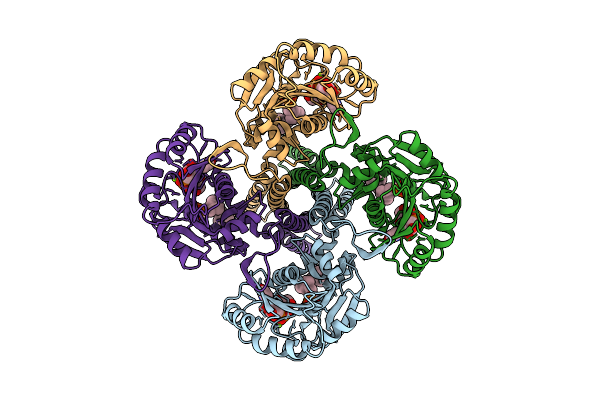 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Substrate-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
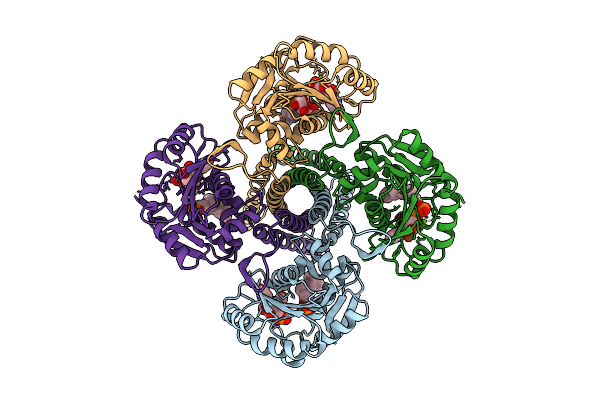 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Catalysis And Product-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG, UDP, A1B94 |
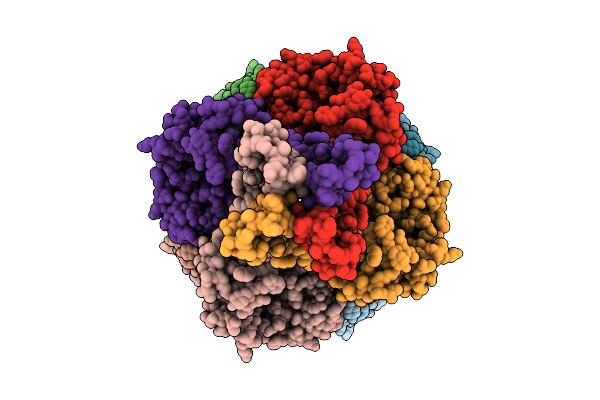 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Apo State (Octamer Volume)
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
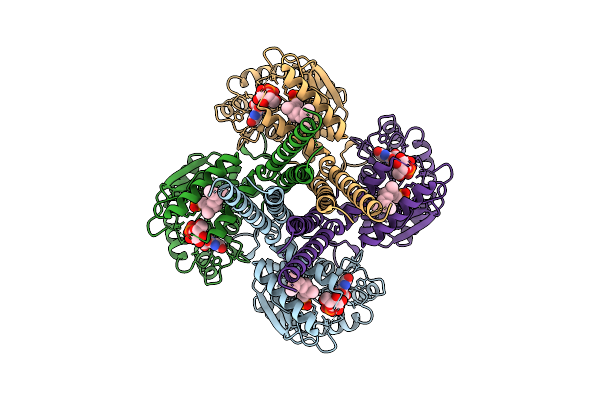 |
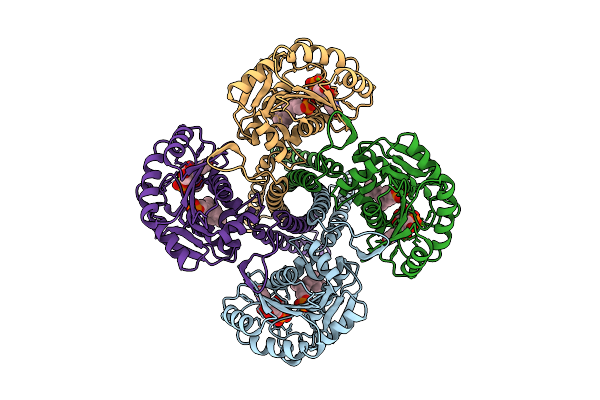
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Intermediate State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
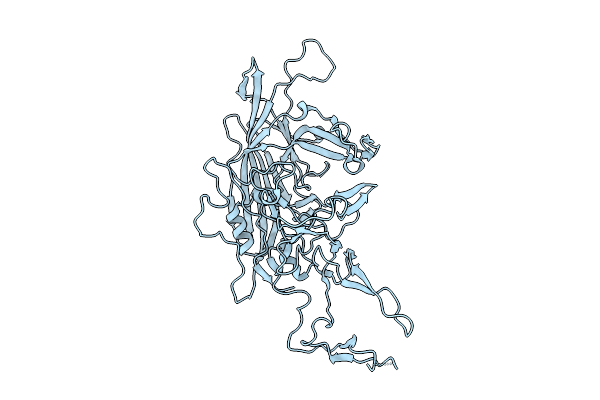 |
Organism: Adeno-associated virus 9
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRUS LIKE PARTICLE |
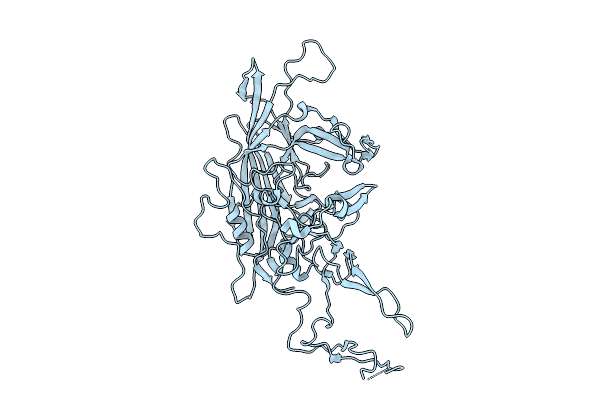 |
Organism: Adeno-associated virus 9
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRUS LIKE PARTICLE |
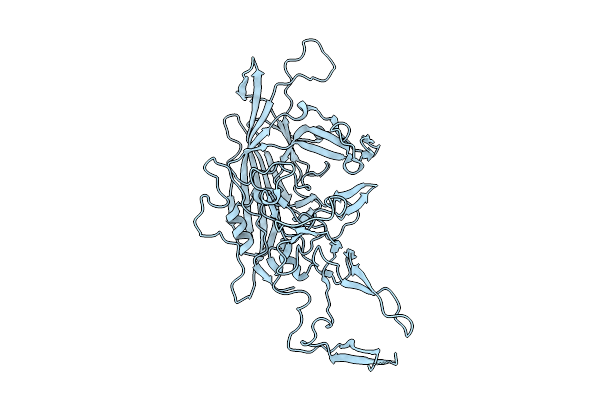 |
Organism: Adeno-associated virus 9
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of The Glycosyltransferase Arnc From Salmonella Enterica In The Apo State Determined On Talos Arctica Microscope
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of The Glycosyltransferase Arnc From Salmonella Enterica In The Udp-Bound State Determined On Talos Arctica Microscope
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: UDP, MN |
 |
Cryo-Em Structure Of The Glycosyltransferase Arnc From Salmonella Enterica In The Apo State Determined On Krios Microscope
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. lt2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSFERASE |
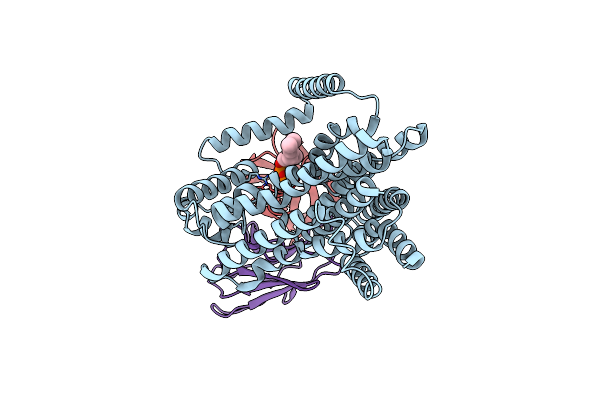 |
Single-Particle Cryo-Em Structure Of The Waal O-Antigen Ligase In Its Ligand Bound State
Organism: Cupriavidus metallidurans, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN Ligands: GPP |
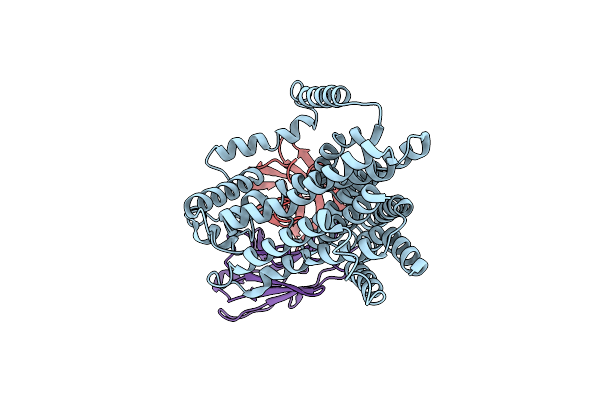 |
Single-Particle Cryo-Em Structure Of The Waal O-Antigen Ligase In Its Apo State
Organism: Cupriavidus metallidurans, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: MEMBRANE PROTEIN |
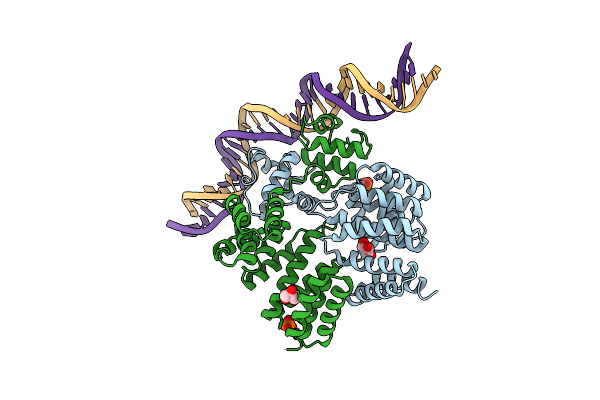 |
Crystal Structure Of Streptococcus Dysgalactiae Shp Pheromone Receptor Rgg2 Bound To Dna
Organism: Streptococcus dysgalactiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-10-21 Classification: DNA BINDING PROTEIN/DNA Ligands: PO4, GOL |
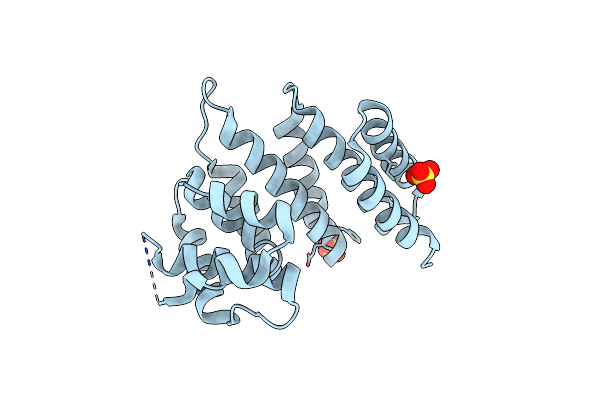 |
Crystal Structure Of Streptococcus Thermophilus Shp Pheromone Receptor Rgg3
Organism: Streptococcus thermophilus (strain atcc baa-250 / lmg 18311)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-10-21 Classification: DNA BINDING PROTEIN Ligands: SO4 |
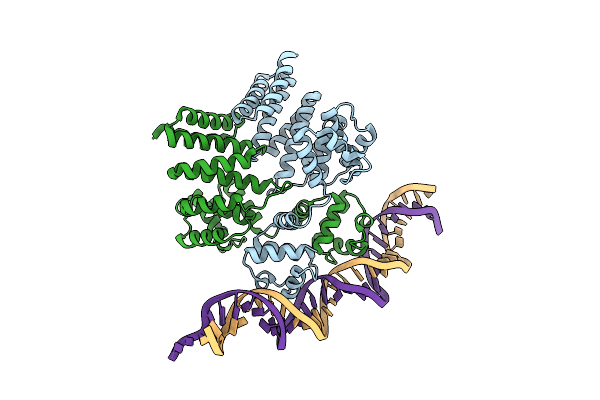 |
Crystal Structure Of Streptococcus Thermophilus Shp Pheromone Receptor Rgg3 Bound To Dna
Organism: Streptococcus thermophilus (strain atcc baa-250 / lmg 18311), Streptococcus thermophilus cnrz1066
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2020-10-21 Classification: DNA BINDING PROTEIN/DNA |
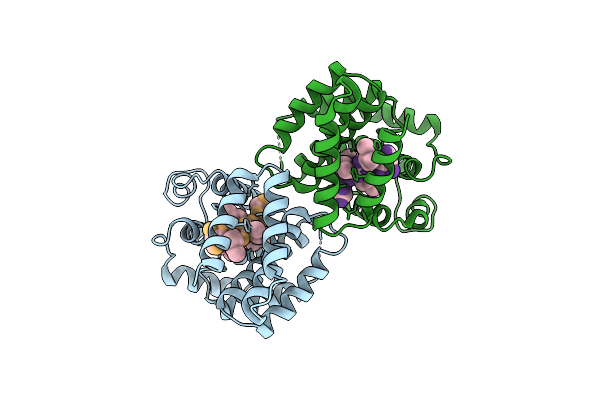 |
Cryoem Structure Of Streptococcus Thermophilus Shp Pheromone Receptor Rgg3 In Complex With Shp3
Organism: Streptococcus thermophilus (strain atcc baa-250 / lmg 18311), Streptococcus thermophilus cnrz1066
Method: ELECTRON MICROSCOPY Release Date: 2020-10-07 Classification: DNA BINDING PROTEIN |
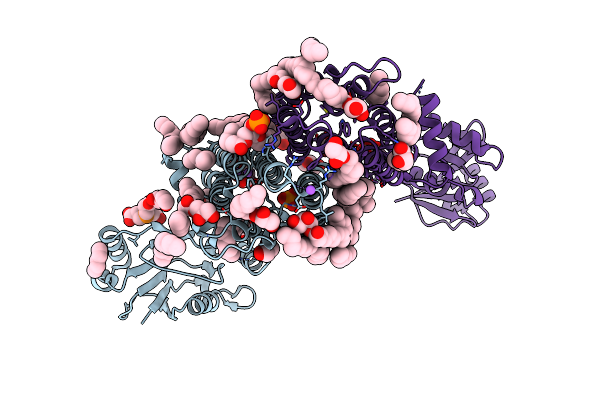 |
Structure Of A Phosphatidylinositol-Phosphate Synthase (Pips) From Mycobacterium Kansasii
Organism: Archaeoglobus fulgidus, Mycobacterium kansasii
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: FLC, NA, PO4, TCE, OLC, 8K6, GOL, XP4 |
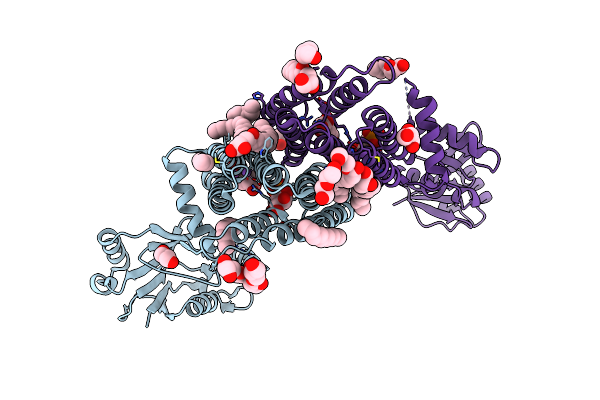 |
Structure Of A Phosphatidylinositol-Phosphate Synthase (Pips) From Mycobacterium Kansasii With Evidence Of Substrate Binding
Organism: Archaeoglobus fulgidus, Mycobacterium kansasii atcc 12478
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: LIP, NA, TCE, 1PE, OLC, 8K6, GOL, C5P, FLC |
 |
Organism: Cupriavidus metallidurans (strain atcc 43123 / dsm 2839 / nbrc 102507 / ch34)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-02-17 Classification: TRANSFERASE Ligands: ZN, DSL, MPG, CL, PO4, PC, EPE |

