Search Count: 14
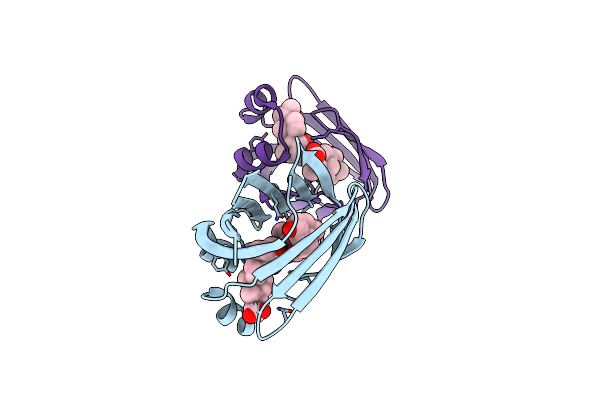 |
Crystal Structure Of Recombinant Chicken Liver Bile Acid Binding Protein (Cl-Babp) In Complex With Chenodeoxycholic Acid
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-10-09 Classification: LIPID BINDING PROTEIN Ligands: JN3 |
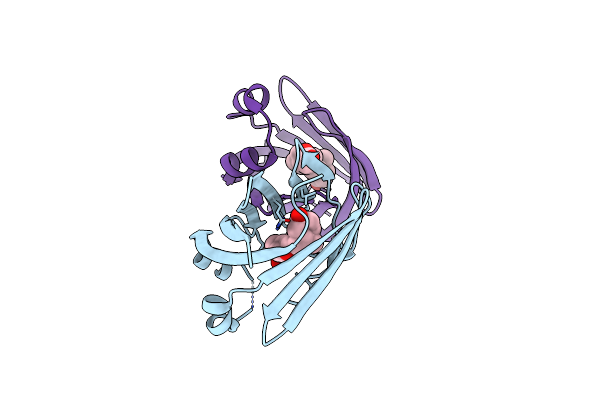 |
Crystal Structure Of Recombinant Chicken Liver Bile Acid Binding Protein (Cl-Babp) In Complex With Ursodeoxycholic Acid
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-10-09 Classification: LIPID BINDING PROTEIN Ligands: IU5 |
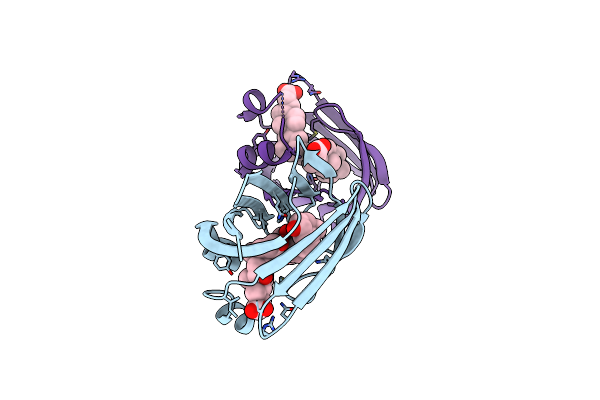 |
Crystal Structure Of Recombinant Chicken Liver Bile Acid Binding Protein (Cl-Babp) In Complex With Deoxycholic Acid
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-10-09 Classification: LIPID BINDING PROTEIN Ligands: DXC |
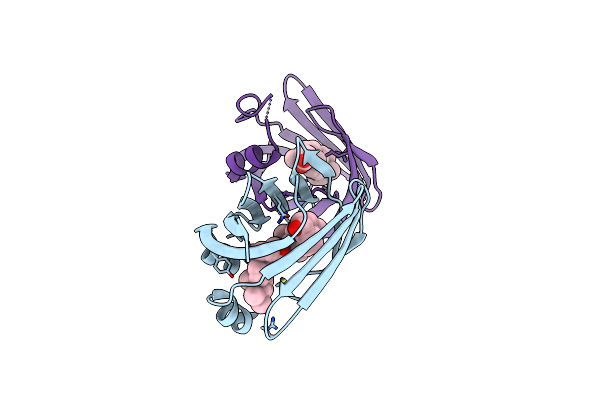 |
Crystal Structure Of Recombinant Chicken Liver Bile Acid Binding Protein (Cl-Babp) In Complex With Lithocholic Acid
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-10-09 Classification: LIPID BINDING PROTEIN Ligands: 4OA |
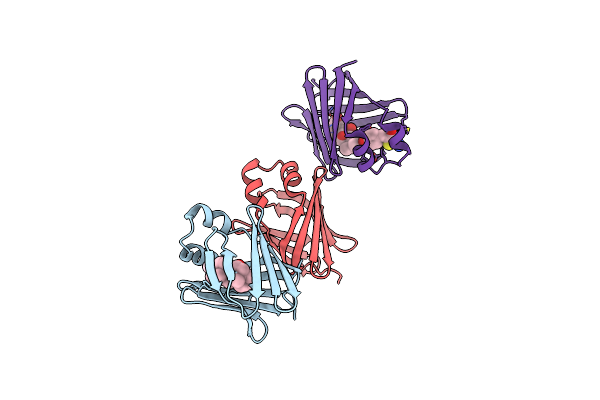 |
Crystal Structure Of Recombinant Chicken Liver Bile Acid Binding Protein (Cl-Babp) In Complex With Ca-M11
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-09 Classification: LIPID BINDING PROTEIN Ligands: A1H7S |
 |
Structure Of Human Heat Shock Protein 90-Alpha N-Terminal Domain (Hsp90-Ntd) In Complex With Jmc31
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-09-07 Classification: CHAPERONE Ligands: M0U |
 |
Structure Of Human Heat Shock Protein 90-Alpha N-Terminal Domain (Hsp90-Ntd) Variant K112A In Complex With Jmc31
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2022-09-07 Classification: CHAPERONE Ligands: M0U |
 |
Structure Of Human Heat Shock Protein 90-Alpha N-Terminal Domain (Hsp90-Ntd) Variant K112R In Complex With Jmc31
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-09-07 Classification: CHAPERONE Ligands: M0U |
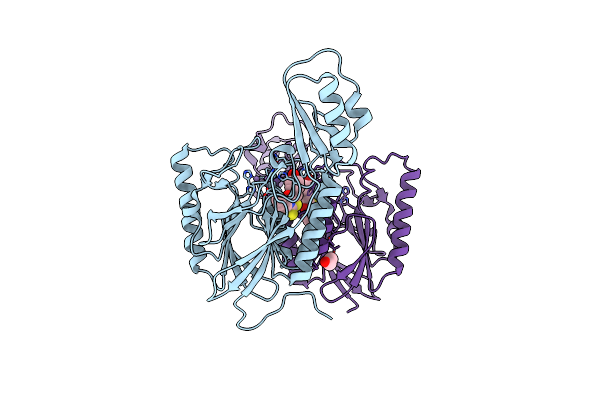 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-03-07 Classification: hydrolase/hydrolase inhibitor Ligands: MN, BU7, EDO |
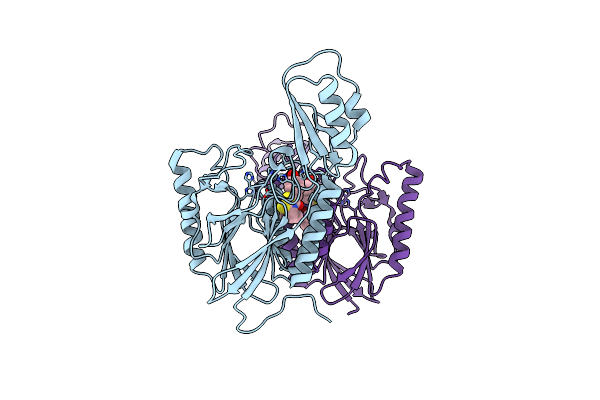 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-01-15 Classification: DNA BINDING PROTEIN/inhibitor Ligands: 2Q0, MN |
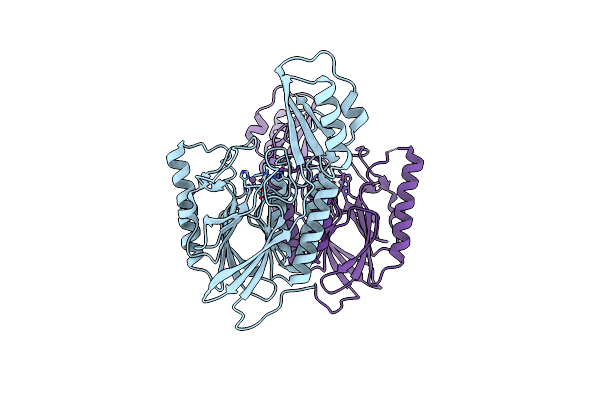 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-01-08 Classification: HYDROLASE Ligands: MN |
 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-01-08 Classification: DNA BINDING PROTEIN/inhibitor Ligands: MN, 2PW |
 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-01-08 Classification: DNA BINDING PROTEIN/inhibitor Ligands: MN, 2PK |
 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-01-08 Classification: DNA BINDING PROTEIN/inhibitor Ligands: MN, 2PV |

