Search Count: 26
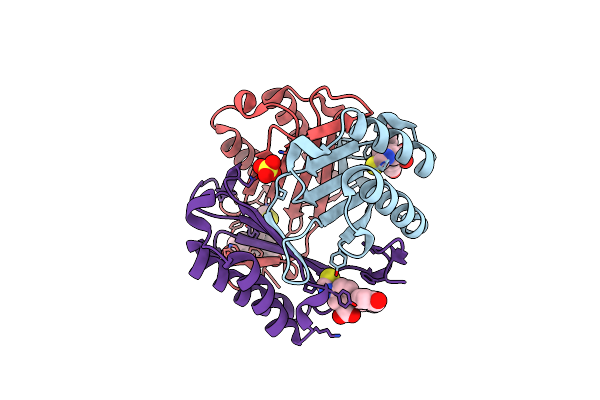 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2018-06-06 Classification: ISOMERASE Ligands: 6B9, SO4 |
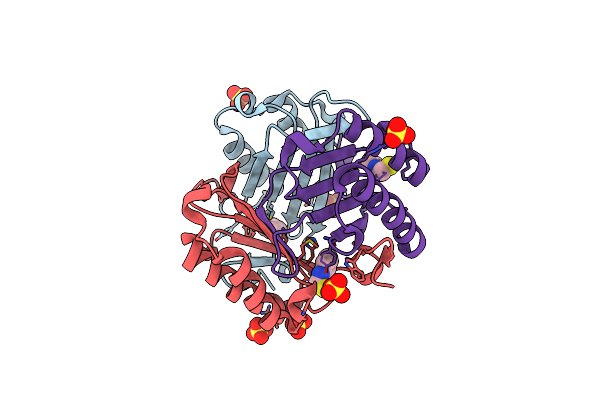 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-06-06 Classification: ISOMERASE Ligands: SO4, 0FI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2018-01-17 Classification: SIGNALING PROTEIN Ligands: NAG |
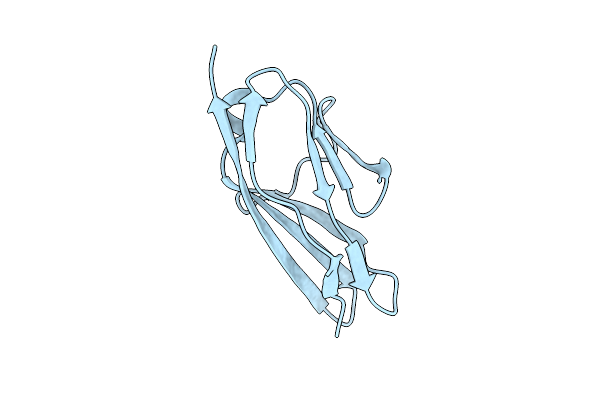 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2018-01-17 Classification: SIGNALING PROTEIN |
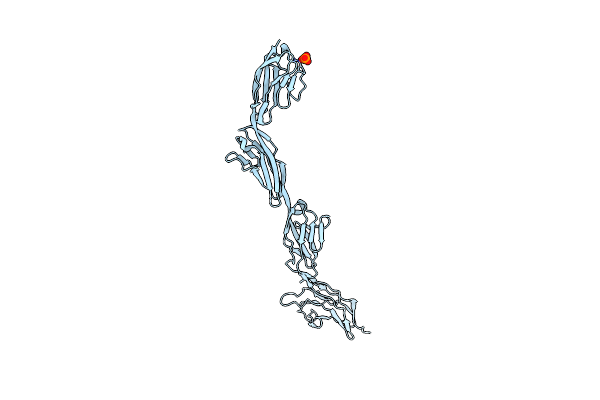 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2018-01-17 Classification: SIGNALING PROTEIN Ligands: PO4 |
 |
Kshv Lana (Orf73) C-Terminal Domain, Open Non-Ring Conformation: Orthorhombic Crystal Form
Organism: Human herpesvirus 8
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2015-10-28 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Crystal Structure Of The Ba-Soaked C2 Crystal Form Of Pmv158 Replication Initiator Repb (P3221 Space Group)
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2015-08-26 Classification: REPLICATION Ligands: CL, MN, BA |
 |
Ca-Bound Truncated (Delta13C) And C3S, C81S And C86S Mutated S100A4 Complexed With Non-Muscle Myosin Iia
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2014-05-07 Classification: CA-BINDING PROTEIN/MOTOR PROTEIN Ligands: CA |
 |
Ca-Bound S100A4 C3S, C81S, C86S And F45W Mutant Complexed With Non- Muscle Myosin Iia
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-05-07 Classification: CA-BINDING PROTEIN/MOTOR PROTEIN Ligands: CA |
 |
Crystal Structure Of E153Q Mutant Of Cold-Adapted Chitinase From Moritella Marina
Organism: Moritella marina
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2014-03-19 Classification: HYDROLASE |
 |
Crystal Structure Of E153Q Mutant Of Cold-Adapted Chitinase From Moritella Complex With Nag4
Organism: Moritella marina
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2014-03-19 Classification: HYDROLASE Ligands: GOL, NA, SO4, GLY |
 |
Crystal Structure Of E153Q Mutant Of Cold-Adapted Chitinase From Moritella Complex With Nag5
Organism: Moritella marina
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2014-03-19 Classification: HYDROLASE |
 |
Crystal Structure Of Full-Length E.Coli Dna Mismatch Repair Protein Muts D835R Mutant In Complex With Gt Mismatched Dna
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-07-17 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-04-17 Classification: OXIDOREDUCTASE Ligands: NAG, CU, O, OXY, CA, NA, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.69 Å Release Date: 2013-04-10 Classification: OXIDOREDUCTASE Ligands: CU, NAG, HEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2009-10-27 Classification: REPLICATION |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2009-07-28 Classification: LYASE |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2008-10-07 Classification: HYDROLASE |
 |
The 9.5 A Resolution Structure Of Glutamate Synthase From Cryo-Electron Microscopy And Its Oligomerization Behavior In Solution: Functional Implications.
Organism: Azospirillum brasilense
Method: ELECTRON MICROSCOPY Resolution:9.50 Å Release Date: 2008-01-15 Classification: OXIDOREDUCTASE Ligands: OMT, FMN, AKG, F3S, SF4, FAD |
 |
Organism: Mason-pfizer monkey virus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2006-11-21 Classification: HYDROLASE |

