Search Count: 12
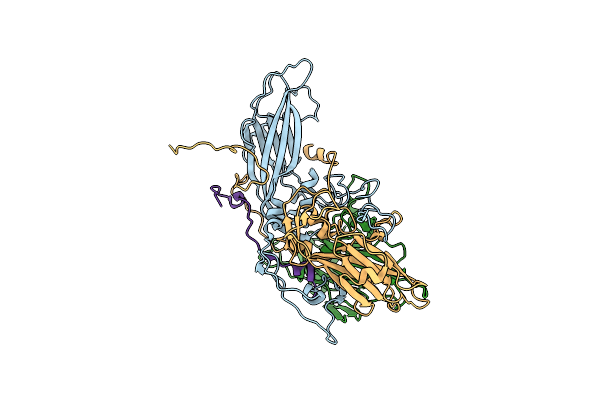 |
Organism: Enterovirus d68
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: VIRUS |
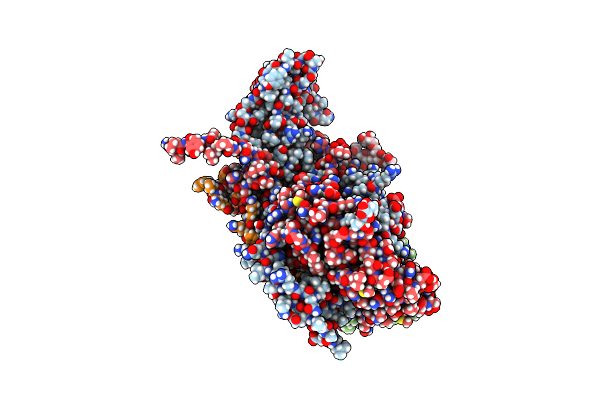 |
Cryo-Em Structure Of Human Enterovirus D68 Usa/Il/14-18952 In Complex With Fc-Mfsd6(L3)
Organism: Homo sapiens, Enterovirus d68
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: VIRUS |
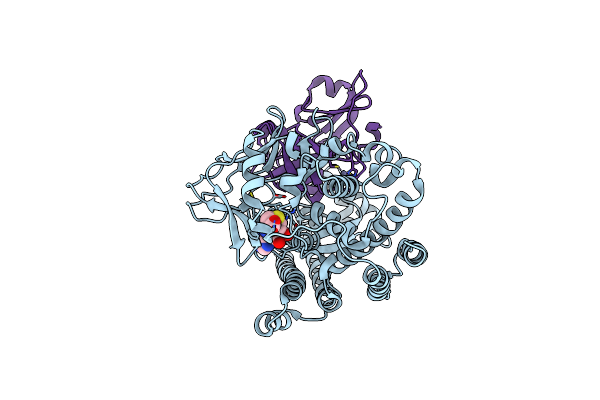 |
Structure Of Human Setd3 Methyl-Transferase In Complex With 2A Protease From Coxsackievirus B3
Organism: Coxsackievirus b3 (strain nancy), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: VIRAL PROTEIN Ligands: ZN, SAH |
 |
Pseudo-Atomic Model For Hsp26 Residues 63 To 214. Please Be Advised That The Target Map Is Not Of Sufficient Resolution To Unambiguously Position Backbone Or Side Chain Atoms. This Model Represents A Likely Fit.
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-11-24 Classification: CHAPERONE |
 |
Pseudo-Atomic Model Of A 16-Mer Assembly Of Reduced Recombinant Human Alphaa-Crystallin (Non Domain Swapped Configuration)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-12-11 Classification: CHAPERONE |
 |
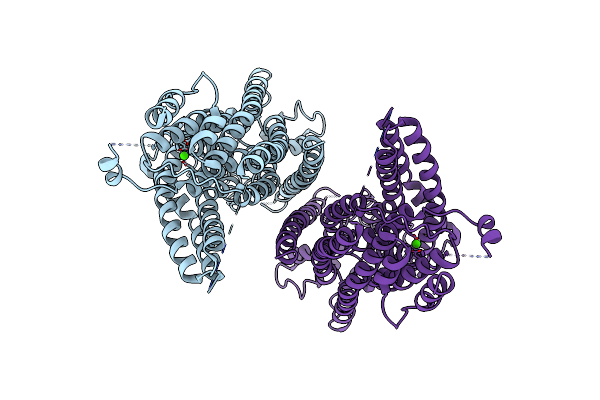
Cryo-Em Structure Of The Tmem16A Calcium-Activated Chloride Channel In Nanodisc
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-12-27 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Cryo-Em Structure Of The Tmem16A Calcium-Activated Chloride Channel In Lmng
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-12-27 Classification: MEMBRANE PROTEIN Ligands: CA |
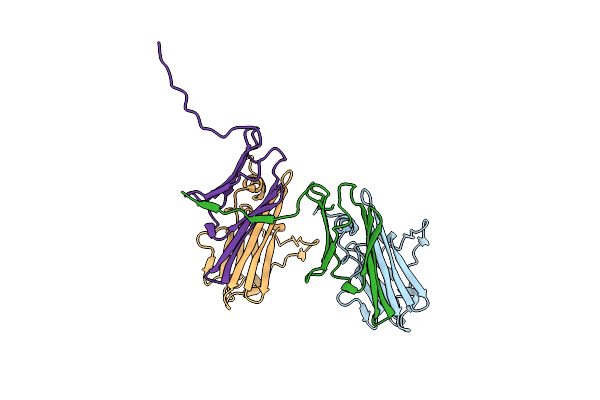 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2015-06-10 Classification: CHAPERONE |
 |
Stress-Induced Protein 1 Truncation Mutant (43 - 140) From Caenorhabditis Elegans
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-06-10 Classification: CHAPERONE Ligands: SO4 |
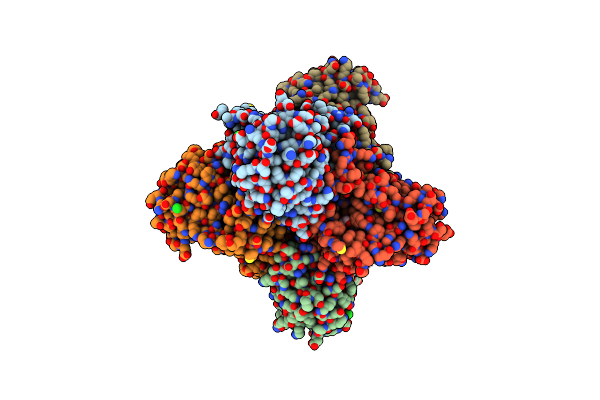 |
Bacterial Chalcone Isomerase In Closed Conformation From Eubacterium Ramulus At 2.8 A Resolution
Organism: Eubacterium ramulus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-01-15 Classification: ISOMERASE Ligands: GOL, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-11-10 Classification: MOTOR PROTEIN Ligands: MG, ADP |
 |
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2007-05-08 Classification: HYDROLASE Ligands: NOL |

