Search Count: 87
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSCRIPTION/INHIBITOR Ligands: BME, 9N6, NA, GOL, SO4 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH4 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH5 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH6 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH7 |
 |
Cyclohexanone Dehydrogenase (Cdh) From Alicycliphilus Denitrificans K601 - Wildtype
Organism: Alicycliphilus denitrificans k601
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-02-14 Classification: FLAVOPROTEIN Ligands: FAD, GOL, SO4 |
 |
Cyclohexanone Dehydrogenase (Cdh) From Alicycliphilus Denitrificans K601 Complexed With Dehydrogenated Substrate Cyclohex-2-En-1-One - Inactive Mutant (Y195F)
Organism: Alicycliphilus denitrificans k601
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2024-02-14 Classification: FLAVOPROTEIN Ligands: FAD, A2Q, GOL, SO4 |
 |
Cyclohexanone Dehydrogenase (Cdh) From Alicycliphilus Denitrificans K601 Complexed With Dehydrogenated Substrate - W113A Mutant
Organism: Alicycliphilus denitrificans k601
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-02-14 Classification: FLAVOPROTEIN Ligands: FAD, GOL, PEG, A2Q, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: CL, ZQW |
 |
Co-Crystal Structure Of Capcna Bound To The Aoh1996 Derivative, Aoh1996-1Le
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.77 Å Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: ZQZ, CL |
 |
Sars-Cov-2 Replication-Transcription Complex Bound To Remdesivir Triphosphate, In A Pre-Catalytic State
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: REPLICATION Ligands: NWX, MG, ZN |
 |
Sars-Cov-2 Replication-Transcription Complex Bound To Atp, In A Pre-Catalytic State
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: REPLICATION Ligands: ATP, MG, ZN |
 |
Sars-Cov-2 Replication-Transcription Complex Bound To Utp, In A Pre-Catalytic State
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: REPLICATION Ligands: ZN, MG, UTP |
 |
Sars-Cov-2 Replication-Transcription Complex Bound To Gtp, In A Pre-Catalytic State
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: REPLICATION Ligands: ZN, MG, GTP, L2B |
 |
Sars-Cov-2 Replication-Transcription Complex Bound To Ctp, In A Pre-Catalytic State
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: REPLICATION Ligands: ZN, MG, CTP, L2B |
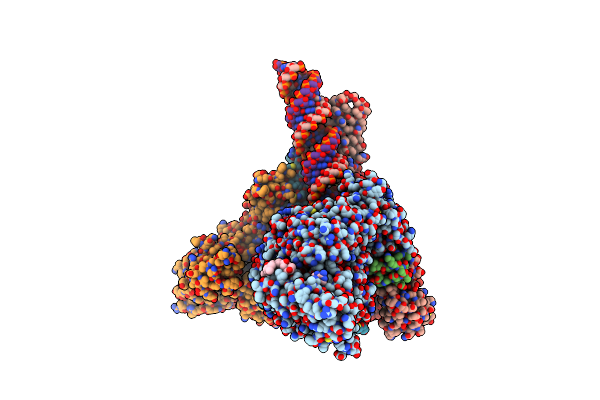 |
Sars-Cov-2 Replication-Transcription Complex Bound To Nsp13 Helicase - Nsp13(2)-Rtc - Engaged Class
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: REPLICATION/TRANSCRIPTION Ligands: ZN, MG, ADP, 1N7, AF3 |
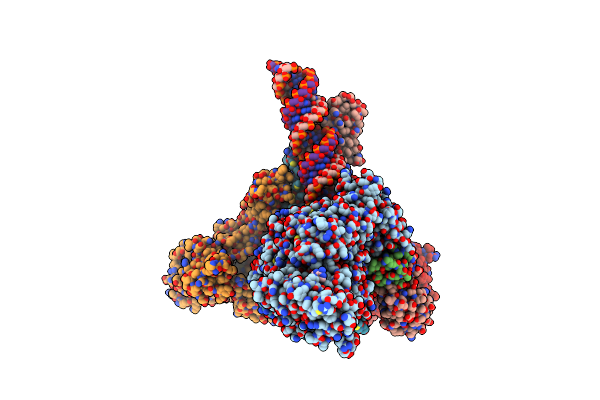 |
Sars-Cov-2 Replication-Transcription Complex Bound To Nsp13 Helicase - Nsp13(2)-Rtc - Swiveled Class
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: REPLICATION/TRANSCRIPTION Ligands: ZN, MG, ADP, AF3 |
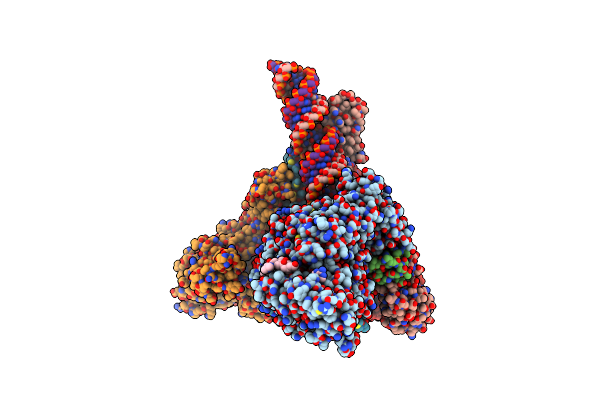 |
Sars-Cov-2 Replication-Transcription Complex Bound To Nsp13 Helicase - Nsp13(2)-Rtc (Composite)
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: REPLICATION/TRANSCRIPTION Ligands: ZN, MG, ADP, 1N7, AF3 |
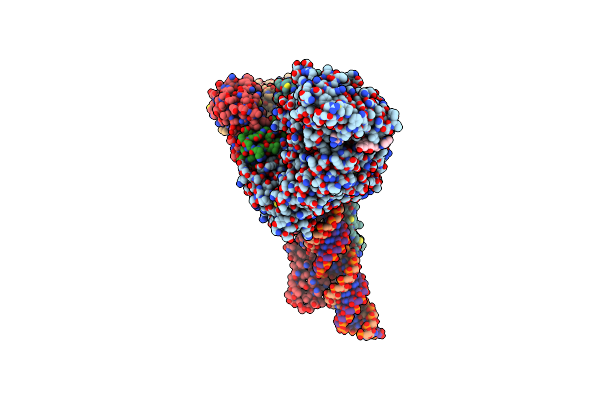 |
Sars-Cov-2 Replication-Transcription Complex Bound To Nsp13 Helicase - Nsp13(1)-Rtc
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: REPLICATION/TRANSCRIPTION Ligands: ZN, MG, ADP, 1N7, AF3 |
 |
Sars-Cov-2 Replication-Transcription Complex Bound To Nsp13 Helicase - Nsp13(2)-Rtc - Open Class
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-11-24 Classification: REPLICATION/TRANSCRIPTION Ligands: ZN, MG, ADP, 1N7, AF3 |

