Search Count: 22
 |
The Open Conformation Of E.Coli Elongation Factor Tu In Complex With Gdpnp.
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2018-08-22 Classification: TRANSLATION Ligands: GNP, MG, GOL, SO4, PEG |
 |
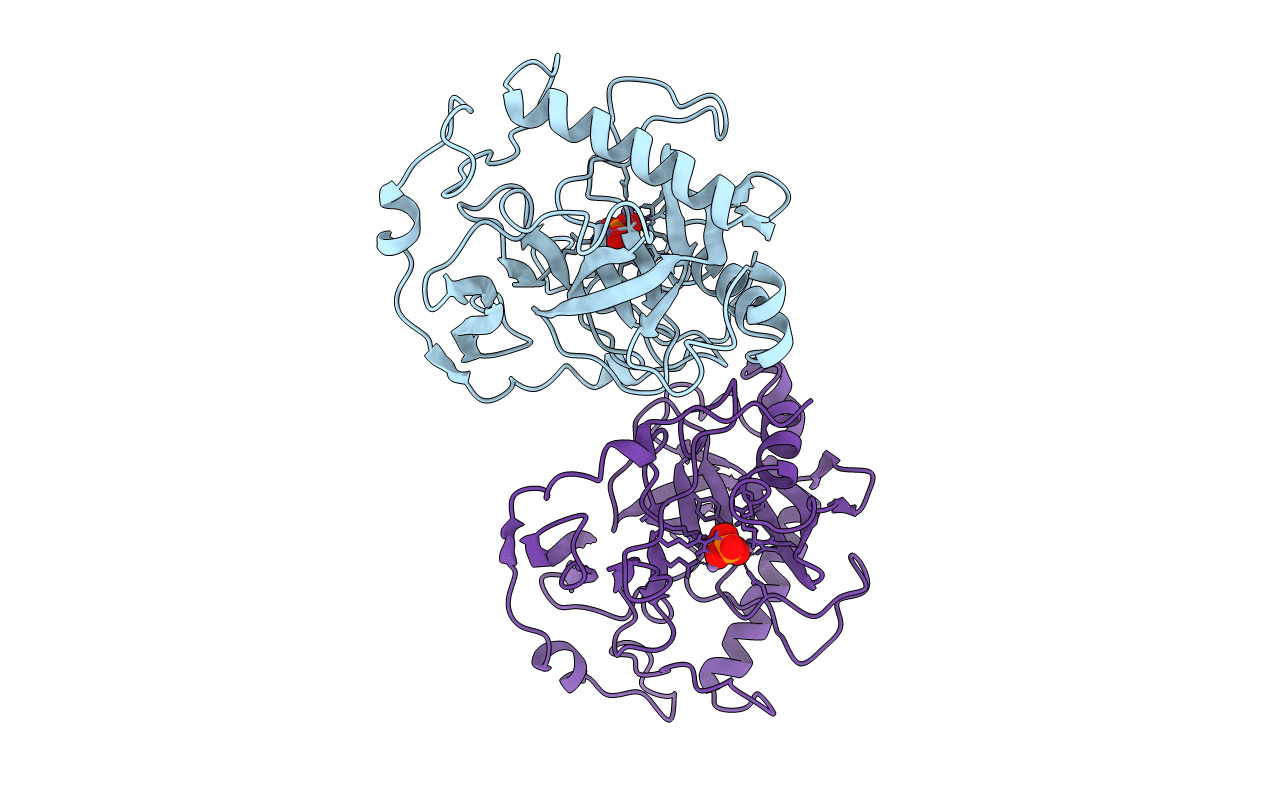
The Structural Basis For Peptidomimetic Inhibition Of Eukaryotic Ribonucleotide Reductase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2008-08-19 Classification: OXIDOREDUCTASE |
 |
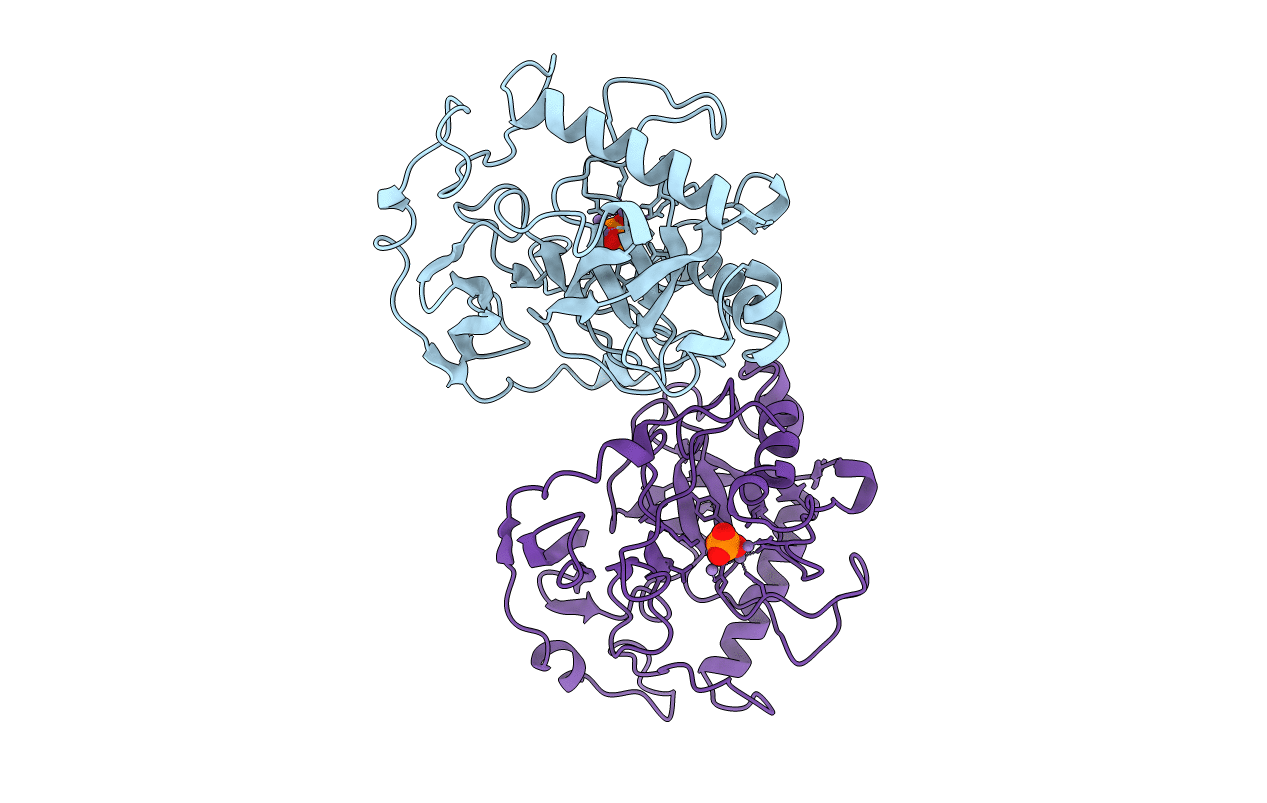
The Structual Basis For Peptidomimetic Inhibition Of Eukaryotic Ribonucleotide Reductase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2008-08-19 Classification: OXIDOREDUCTASE Ligands: MRT, GOL |
 |
Structural Basis For Ligand Binding And Heparin Mediated Activation Of Neuropilin
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-04-03 Classification: SIGNALING PROTEIN, MEMBRANE PROTEIN |
 |
Structural Basis For Ligand Binding And Heparin Mediated Activation Of Neuropilin
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2007-04-03 Classification: SIGNALING PROTEIN, MEMBRANE PROTEIN |
 |
Crystal Structures Of Eap Domains From Staphylococcus Aureus Reveal An Unexpected Homology To Bacterial Superantigens
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2005-03-01 Classification: TOXIN, PROTEIN BINDING |
 |
Crystal Structures Of Eap Domains From Staphylococcus Aureus Reveal An Unexpected Homology To Bacterial Superantigens
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-03-01 Classification: UNKNOWN FUNCTION Ligands: ZN |
 |
Crystal Structures Of Eap Domains From Staphylococcus Aureus Reveal An Unexpected Homology To Bacterial Superantigens
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-03-01 Classification: UNKNOWN FUNCTION |
 |
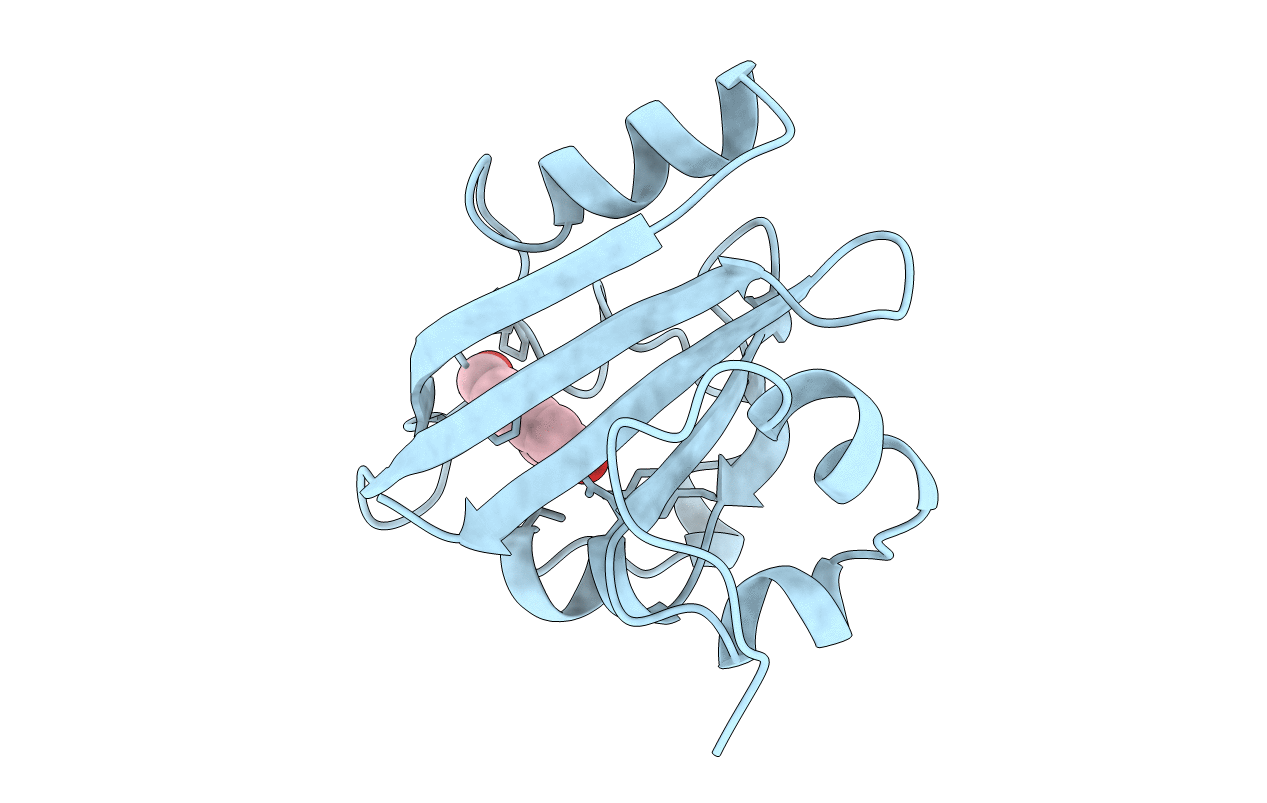
Fluoride-Inhibited Substrate Complex Of Saccharomyces Cerevisiae Inorganic Pyrophosphatase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2001-03-19 Classification: PHOSPHORYL TRANSFER Ligands: MN, POP, F, NA, PO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2001-03-19 Classification: HYDROLASE Ligands: MN, PO4 |
 |
Structure Of Cyclic Peptide Inhibitors Of Mammalian Ribonucleotide Reductase
|
 |
Organism: Halorhodospira halophila
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1999-04-06 Classification: PHOTORECEPTOR Ligands: HC4 |
 |
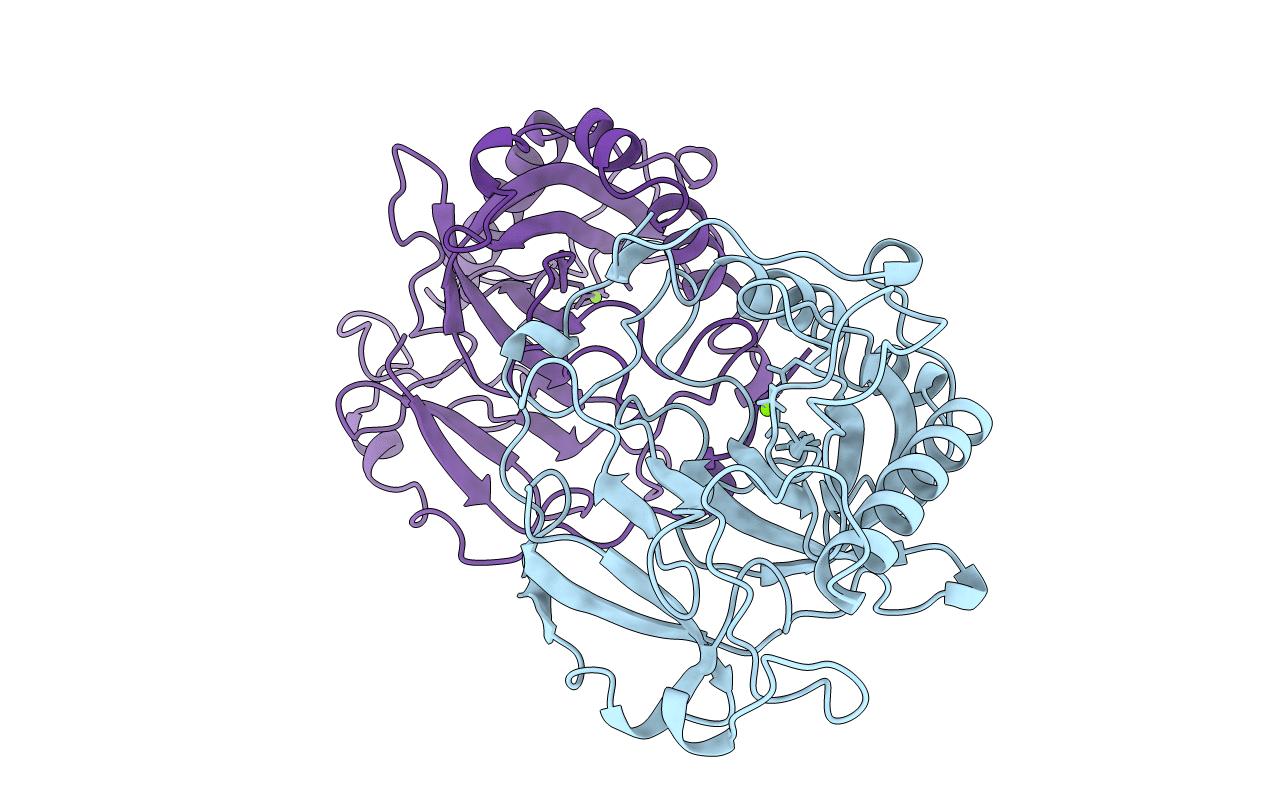
The R78K And D117E Active Site Variants Of Saccharomyces Cerevisiae Soluble Inorganic Pyrophosphatase: Structural Studies And Mechanistic Implications
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 1998-12-23 Classification: HYDROLASE Ligands: MN, PO4 |
 |
The R78K And D117E Active Site Variants Of Saccharomyces Cerevisiae Soluble Inorganic Pyrophosphatase: Structural Studies And Mechanistic Implications
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1998-12-23 Classification: HYDROLASE Ligands: MN, PO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1998-04-08 Classification: HYDROLASE Ligands: MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1998-04-08 Classification: HYDROLASE Ligands: MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1997-11-19 Classification: HYDROLASE Ligands: MN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1997-11-19 Classification: HYDROLASE Ligands: MN, PO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1997-08-20 Classification: HYDROLASE Ligands: MG |
 |
Small Subunit C-Terminal Inhibitory Peptide Of Mouse Ribonucleotide Reductase As Bound To The Large Subunit, Nmr, 26 Structures
|

