Search Count: 30
 |
Deletions In A Surface Loop Divert The Folding Of A Protein Domain Into A Metastable Dimeric Form
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2009-09-15 Classification: TRANSFERASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-02-27 Classification: OXIDOREDUCTASE Ligands: AMP |
 |
Structure Of Protein Ta0514, Putative Lipoate Protein Ligase From T. Acidophilum.
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-12-15 Classification: LIGASE |
 |
Structure Of Protein Ta0514, Putative Lipoate Protein Ligase From T. Acidophilum With Bound Lipoic Acid
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2005-12-15 Classification: LIGASE Ligands: LPA |
 |
New Crystal Form Of The Pseudomonas Putida Branched-Chain Dehydrogenase (E1)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2005-08-10 Classification: OXIDOREDUCTASE |
 |
The Crystal Structure Of Pyruvate Dehydrogenase E1 Bound To The Peripheral Subunit Binding Domain Of E2
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-11-02 Classification: OXIDOREDUCTASE Ligands: MG, PEG, TPP, K |
 |
The Crystal Structure Of Pyruvate Dehydrogenase E1(D180N,E183Q) Bound To The Peripheral Subunit Binding Domain Of E2
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-11-02 Classification: OXIDOREDUCTASE Ligands: MG, TPP |
 |
Nmr Structure Of The Peripheral-Subunit Binding Domain Of Bacillus Stearothermophilus E2P
Organism: Geobacillus stearothermophilus
Method: SOLUTION NMR Release Date: 2004-07-20 Classification: TRANSFERASE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2002-05-29 Classification: PROTEIN TRANSPORT Ligands: ZN, TRS |
 |
Solution Structure Of The Lipoyl Domain Of The Chimeric Dihydrolipoyl Dehydrogenase P64K From Neisseria Meningitidis
Organism: Neisseria meningitidis
Method: SOLUTION NMR Release Date: 2001-11-28 Classification: OXIDOREDUCTASE |
 |
Organism: Bacteriophage ph75
Method: FIBER DIFFRACTION Resolution:2.40 Å Release Date: 2001-06-01 Classification: VIRUS |
 |
Organism: Bacteriophage ph75
Method: FIBER DIFFRACTION Resolution:2.40 Å Release Date: 2001-06-01 Classification: VIRUS |
 |
Organism: Bacteriophage ph75
Method: FIBER DIFFRACTION Resolution:2.40 Å Release Date: 2001-06-01 Classification: VIRUS |
 |
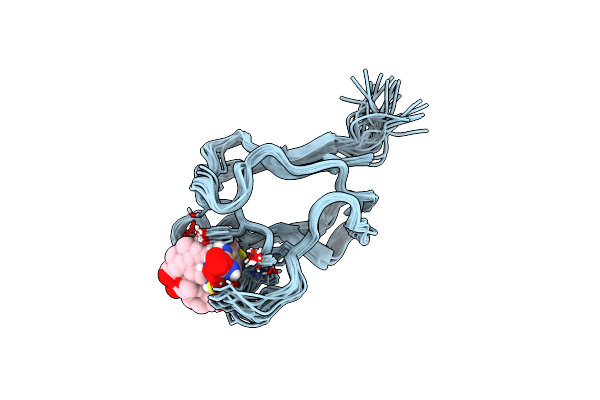
Innermost Lipoyl Domain Of The Pyruvate Dehydrogenase From Escherichia Coli
Organism: Escherichia coli bl21(de3)
Method: SOLUTION NMR Release Date: 1999-06-30 Classification: DIHYDROLIPOAMIDE ACETYLTRANSFERASE |
 |
Solution Structure Of Holo-Biotinyl Domain From Acetyl Coenzyme A Carboxylase Of Escherichia Coli Determined By Triple-Resonance Nmr Spectroscopy
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 1999-04-27 Classification: BIOTIN Ligands: BTN |
 |
Solution Structure Of Apo-Biotinyl Domain From Acetyl Coenzyme A Carboxylase Of Escherichia Coli Determined By Triple-Resonance Nmr Spectroscopy
|
 |
Dihydrolipoyl Transacetylase (E.C.2.3.1.12) Catalytic Domain (Residues 184-425) From Bacillus Stearothermophilus
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:4.40 Å Release Date: 1999-02-16 Classification: ACYLTRANSFERASE |
 |
Inovirus (Filamentous Bacteriophage) Strain Pf3 Major Coat Protein Assembly
Organism: Pseudomonas phage pf3
Method: FIBER DIFFRACTION Resolution:3.10 Å Release Date: 1998-11-04 Classification: VIRUS |
 |
Lipoyl Domain From The Dihydrolipoyl Succinyltransferase Component Of The 2-Oxoglutarate Dehydrogenase Multienzyme Complex Of Escherichia Coli, Nmr, 25 Structures
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 1998-07-29 Classification: TRANSFERASE |
 |
Inner Lipoyl Domain From Human Pyruvate Dehydrogenase (Pdh) Complex, Nmr, 1 Structure
|

