Search Count: 27
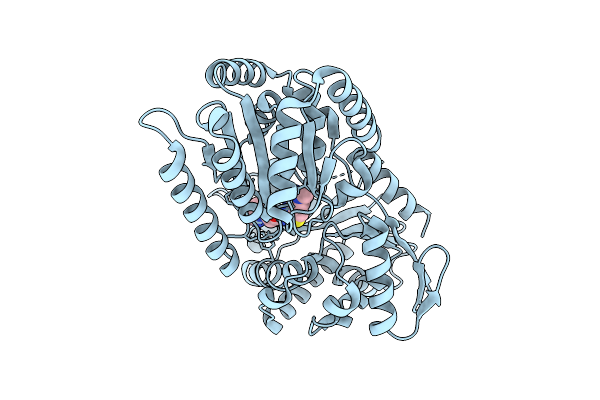 |
Structure Of Candida Albicans Trehalose-6-Phosphate Synthase In Complex With 4456
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: A1BYI |
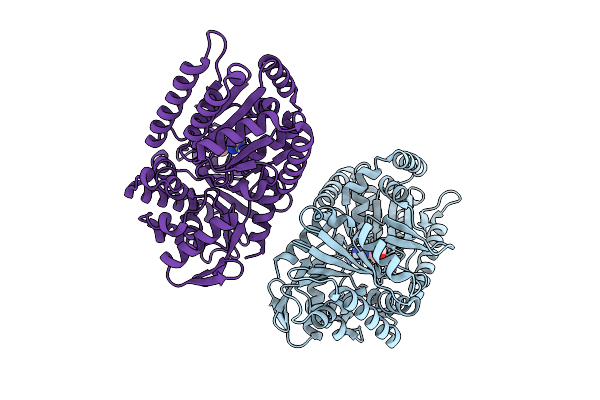 |
Structure Of Candida Albicans Trehalose-6-Phosphate Synthase In Complex With Sj6675
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: A1BYV |
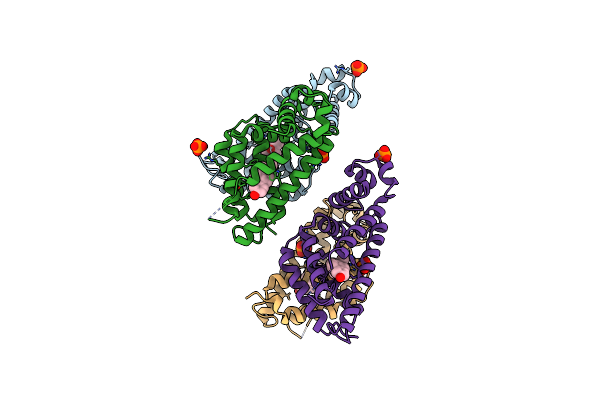 |
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-02-14 Classification: TRANSCRIPTION Ligands: 3WF, PO4 |
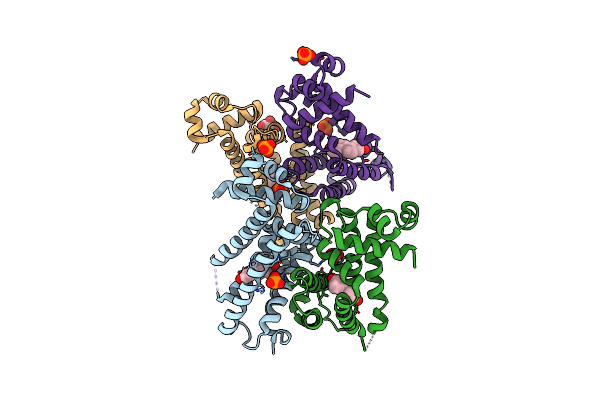 |
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-01-24 Classification: TRANSCRIPTION Ligands: EST, PO4, GOL |
 |
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2024-01-24 Classification: TRANSCRIPTION Ligands: TES, PO4 |
 |
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2024-01-24 Classification: TRANSCRIPTION Ligands: CXS, PO4, STR |
 |
Cryo-Em Structure Of Cryptococcus Neoformans Trehalose-6-Phosphate Synthase Homotetramer In Complex With Uridine Diphosphate And Glucose-6-Phosphate
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: TRANSFERASE Ligands: UDP, G6P |
 |
Organism: Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2021-04-14 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2021-03-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Dh898.1 Fab-Dimer Bound Near The Cd4 Binding Site Of Hiv-1 Env Ch848 Sosip Trimer
Organism: Human immunodeficiency virus 1, Simian-human immunodeficiency virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Dh898.1 Fab-Dimer From Local Refinement Of The Fab-Dimer Bound Near The Cd4 Binding Site Of Hiv-1 Env Ch848 Sosip Trimer
Organism: Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2021-02-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2020-12-30 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
Crystal Structure Of The Disulfide Linked Dh717.1 Fab Dimer, Derived From A Macaque Hiv-1 Vaccine-Induced Env Glycan-Reactive Neutralizing Antibody B Cell Lineage
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2020-12-30 Classification: IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 2P S Ectodomain Bound To One Copy Of Domain-Swapped Antibody 2G12
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 2P S Ectodomain Bound To Two Copies Of Domain-Swapped Antibody 2G12
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 2P S Ectodomain Bound Domain-Swapped Antibody 2G12 From Masked 3D Refinement
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Structure Of Candida Albicans Trehalose-6-Phosphate Synthase In Complex With Udp-Glucose
Organism: Candida albicans (strain sc5314 / atcc mya-2876)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-05-03 Classification: TRANSFERASE Ligands: UPG, SO4, 1PE, NA |
 |
Structure Of Candida Albicans Trehalose-6-Phosphate Synthase In Complex With Udp And Glucose-6-Phosphate
Organism: Candida albicans (strain sc5314 / atcc mya-2876)
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2017-05-03 Classification: TRANSFERASE Ligands: UDP, G6P |
 |
Structure Of Candida Albicans Trehalose-6-Phosphate Synthase In Complex With Udp And Validoxylamine A
Organism: Candida albicans (strain sc5314 / atcc mya-2876)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-05-03 Classification: TRANSFERASE Ligands: UDP, VDM, SO4 |
 |
Structure Of Aspergillus Fumigatus Trehalose-6-Phosphate Synthase A In Complex With Udp And Validoxylamine A
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2017-05-03 Classification: TRANSFERASE Ligands: UDP, VDM |

