Search Count: 57
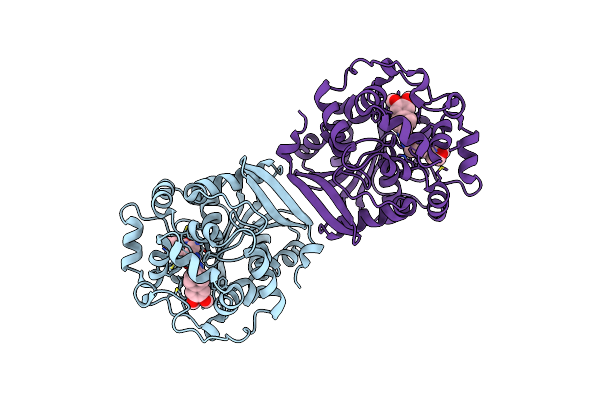 |
Crystal Structure Of Human Soluble Epoxide Hydrolase C-Terminal Domain In Complex With A Benzohomoadamantane-Based Urea Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: HYDROLASE Ligands: A1H8Y |
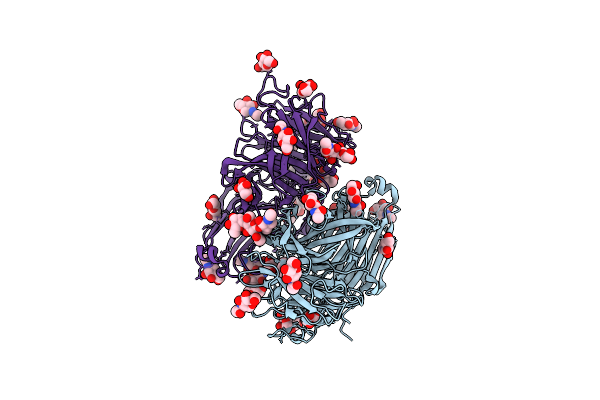 |
Organism: Rhodotorula dairenensis
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: MAN, NAG, PEG, GOL, BTB |
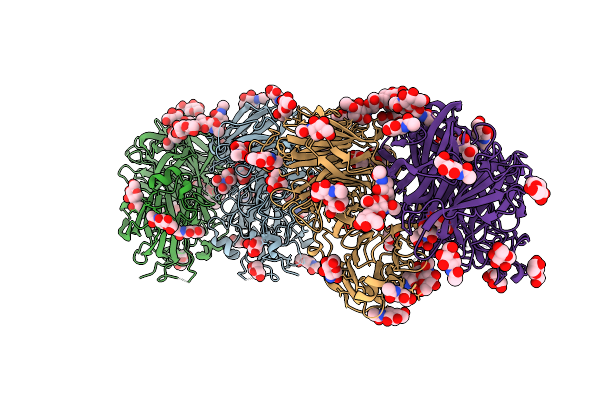 |
Structure Of D188A-Fructofuranosidase From Rhodotorula Dairenensis In Complex With Fructose
Organism: Rhodotorula dairenensis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: MAN, NAG, BTB, FRU, PEG |
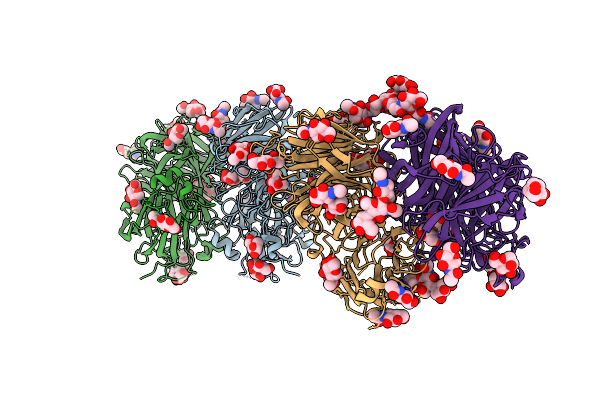 |
Structure Of D188A-Fructofuranosidase From Rhodotorula Dairenesis In Complex With Sucrose
Organism: Rhodotorula dairenensis
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: MAN, NAG |
 |
Structure Of D188A-Fructofuranosidase From Rhodotorula Dairenensis In Complex With Raffinose
Organism: Rhodotorula dairenensis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: MAN, NAG, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-02-02 Classification: ISOMERASE Ligands: MG, NA, CL |
 |
Human Phosphomannomutase 2 (Pmm2) Wild-Type Co-Crystallized With The Activator Glucose 1,6-Bisphosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2022-02-02 Classification: ISOMERASE Ligands: MG, G16, CL |
 |
Human Phosphomannomutase 2 (Pmm2) Wild-Type Soaked With The Activator Glucose 1,6-Bisphosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2022-02-02 Classification: ISOMERASE Ligands: GOL, MG, CL, G16, NA |
 |
Human Phosphomannomutase 2 (Pmm2) With Mutation T237M In Complex With The Activator Glucose 1,6-Bisphosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-02-02 Classification: ISOMERASE Ligands: GOL, G16, MG, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2022-02-02 Classification: ISOMERASE Ligands: GOL, MG, NA, CL |
 |
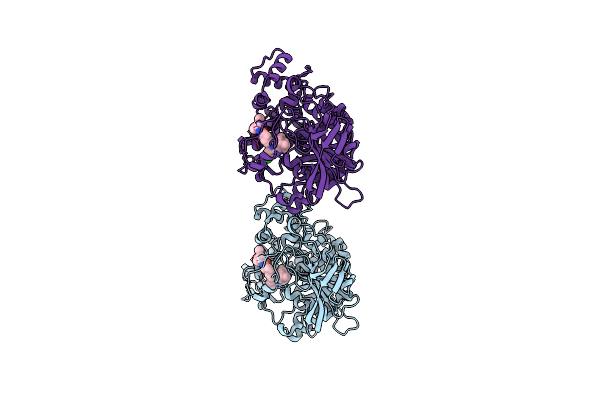
Crystal Structure Of Torpedo Californica Acetylcholinesterase In Complex With 6-[(3-Chloro-6,7,10,11-Tetrahydro-9-Methyl-7,11-Methanocycloocta[B]Quinolin-12-Yl)Amino]-N-(4-Hydroxy-3-Methoxybenzyl)Hexanamide
Organism: Tetronarce californica
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 8UH, NAG, PEG, CL |
 |
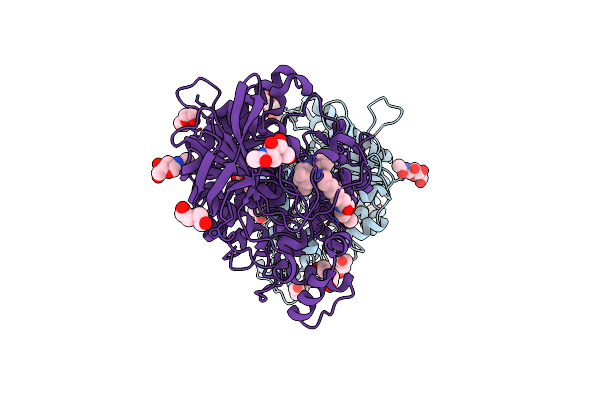
Crystal Structure Of Torpedo Californica Acetylcholinesterase In Complex With 7-[(3-Chloro-6,7,10,11-Tetrahydro-9-Methyl-7,11-Methanocycloocta[B]Quinolin-12-Yl)Amino]-N-(4-Hydroxy-3-Methoxybenzyl)Heptanamide
Organism: Tetronarce californica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 8UK |
 |
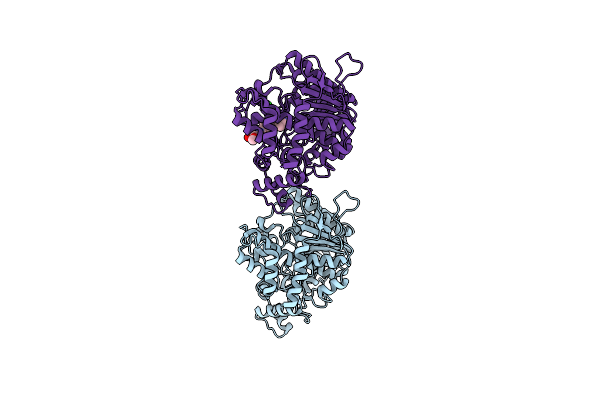
Crystal Structure Of Torpedo Californica Acetylcholinesterase In Complex With 8-[(3-Chloro-6,7,10,11-Tetrahydro-9-Methyl-7,11-Methanocycloocta[B]Quinolin-12-Yl)Amino]-N-(4-Hydroxy-3-Methoxybenzyl)Octanamide
Organism: Tetronarce californica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: PEG, NAG, 8UQ, CL |
 |
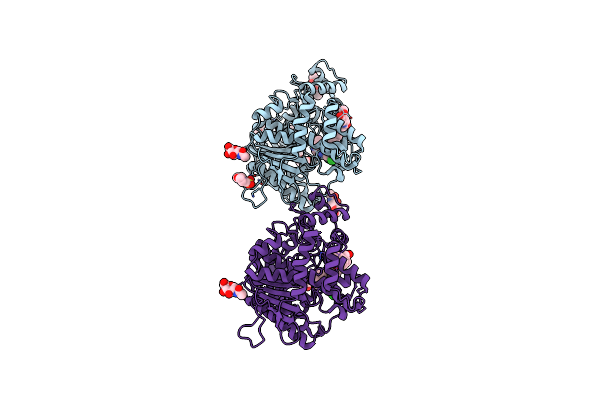
Crystal Structure Of Torpedo Californica Acetylcholinesterase In Complex With 4-{[(3-Chloro-6,7,10,11-Tetrahydro-9-Methyl-7,11-Methanocycloocta[B]Quinolin-12-Yl)Amino]Methyl}-N-(4-Hydroxy-3-Methoxybenzyl)Benzamide
Organism: Tetronarce californica
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 8UE |
 |
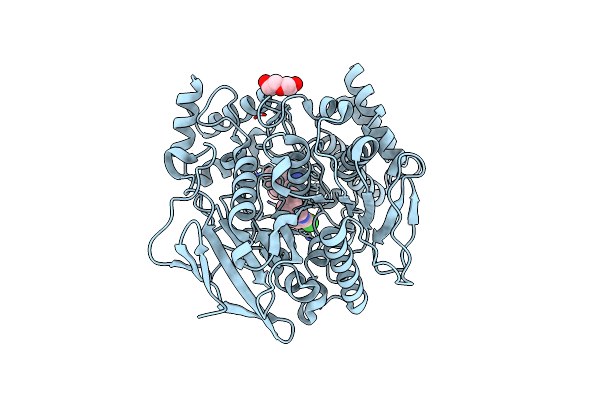
Crystal Structure Of Torpedo Californica Acetylcholinesterase In Complex With (E)-10-[(3-Chloro-6,7,10,11-Tetrahydro-9-Methyl-7,11-Methanocycloocta[B]Quinolin-12-Yl)Amino]-N-(4-Hydroxy-3-Methoxybenzyl)-6-Decenamide
Organism: Tetronarce californica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 8U5, NAG, CL, MES, PEG |
 |
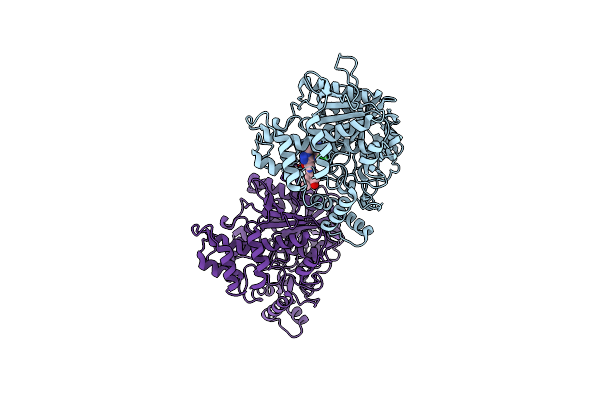
Crystal Structure Of Torpedo Californica Acetylcholinesterase In Complex With 2-{1-[4-(12-Amino-3-Chloro-6,7,10,11-Tetrahydro-7,11-Methanocycloocta[B]Quinolin-9-Yl)Butyl]-1H-1,2,3-Triazol-4-Yl}-N-[4-Hydroxy-3-Methoxybenzyl]Acetamide
Organism: Tetronarce californica
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 8U2, PEG, CL |
 |
Crystal Structure Of Human Butyrylcholinesterase In Complex With 2-{1-[4-(12-Amino-3-Chloro-6,7,10,11-Tetrahydro-7,11-Methanocycloocta[B]Quinolin-9-Yl)Butyl]-1H-1,2,3-Triazol-4-Yl}-N-[4-Hydroxy-3-Methoxybenzyl]Acetamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 8U2 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2021-03-03 Classification: PEPTIDE BINDING PROTEIN Ligands: CL, DMS, RV5 |
 |
Structure Of The Hemiacetal Complex Between The Sars-Cov-2 Main Protease And Leupeptin
Organism: Severe acute respiratory syndrome coronavirus 2, Streptomyces roseus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-03-03 Classification: PEPTIDE BINDING PROTEIN Ligands: DMS, IMD, CL |
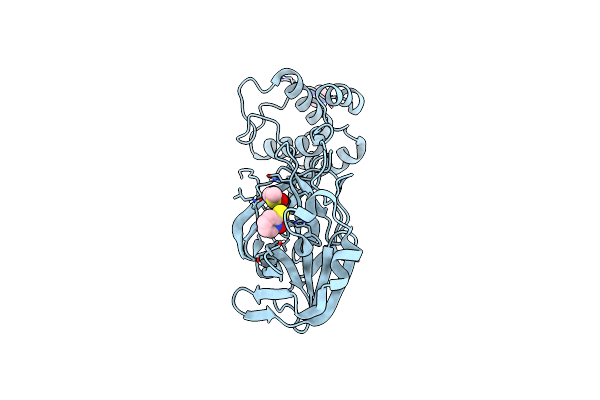 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-01-13 Classification: PEPTIDE BINDING PROTEIN Ligands: PK8, IMD, DMS, CL |

