Search Count: 17
 |
Organism: Cricetulus griseus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-07-07 Classification: HYDROLASE Ligands: AMP, MES, MG |
 |
Organism: Cricetulus griseus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2021-07-07 Classification: HYDROLASE Ligands: AMP, MG, PO4, K, P33, PEG |
 |
Crystal Structure Of Grp78 (70Kda Heat Shock Protein 5 / Bip) Atpase Domain In Complex With Adp And Calcium
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2020-12-16 Classification: CHAPERONE Ligands: ADP, CA |
 |
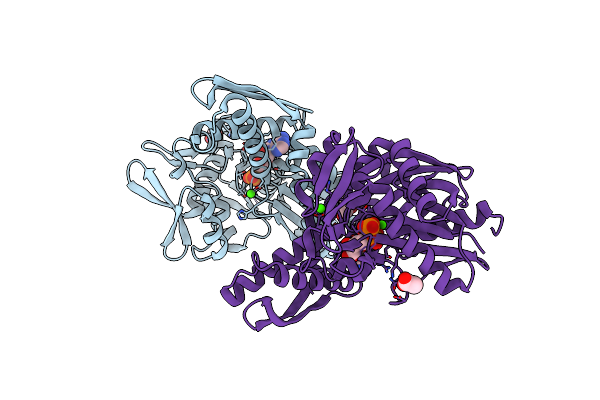
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2020-12-16 Classification: CHAPERONE Ligands: ADP, CA |
 |
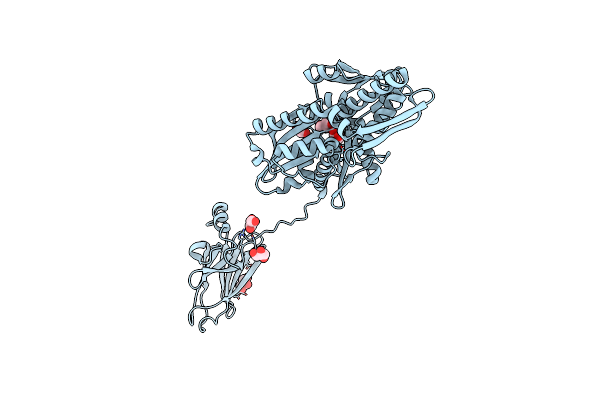
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-12-16 Classification: CHAPERONE Ligands: ACT, CA, ADP, GOL |
 |
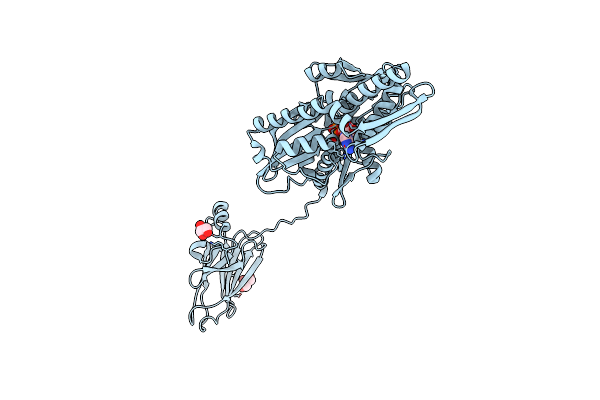
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-12-16 Classification: CHAPERONE Ligands: GOL |
 |
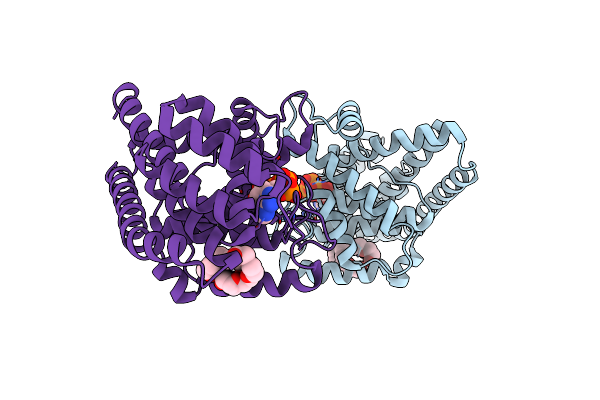
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2020-12-16 Classification: CHAPERONE Ligands: ADP, GOL, K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSLATION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSLATION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSLATION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSLATION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: ATP, PEG, 1PE, PG4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: MG, ATP, P33 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: SO4, 1PE, PG4, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: MG, ATP, EOH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: ANP, MG |

