Search Count: 78
 |
Re-Refined Of Crystal Structure Of Dopa Decarboxylase In Complex With The Inhibitor Carbidopa (1Js3) With Ketoenamine Form Of Carbidopa
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: LYASE Ligands: A1BCS, SO4 |
 |
Cryoem Structure Of Inducible Lysine Decarboxylase From Hafnia Alvei D-Hydrazino-Lysine Analog At 2.3 Angstrom Resolution
Organism: Hafnia alvei atcc 51873
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: LYASE Ligands: A1BD0 |
 |
Cryoem Structure Of Holoenzyme Of Inducible Lysine Decarboxylase From Hafnia Alvei Holoenzyme At 2.19 Angstrom Resolution
Organism: Hafnia alvei atcc 51873
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: LYASE |
 |
Cryoem Structure Of Inducible Lysine Decarboxylase From Hafnia Alvei L-Hydrazino-Lysine Analog At 2.04 Angstrom Resolution
Organism: Hafnia alvei atcc 51873
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: LYASE Ligands: A1BD1 |
 |
X-Ray Structure Of Human Holo Aromatic L-Amino Acid Decarboxylase (Aadc) Complex With Carbidopa At Physiological Ph
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PLP, 142, PG4 |
 |
Human Holo Aromatic L-Amino Acid Decarboxylase (Aadc) R347Q Variant Native Structure
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: LYASE Ligands: PG4 |
 |
Human Holo Aromatic L-Amino Acid Decarboxylase (Aadc) L353P Variant Native Structure
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: LYASE Ligands: PG4 |
 |
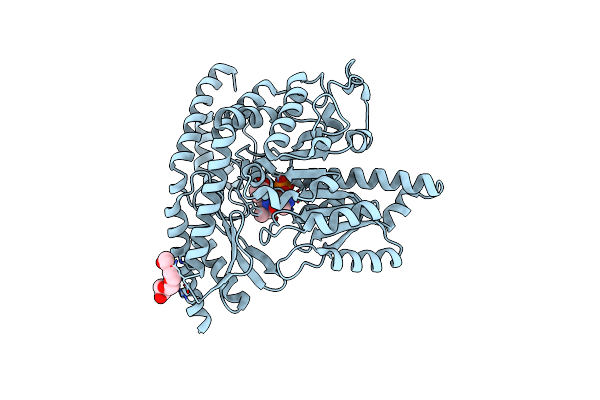
Human Holo Aromatic L-Amino Acid Decarboxylase (Aadc) Native Structure At Physiological Ph
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-08-30 Classification: LYASE Ligands: PLP, PG4 |
 |
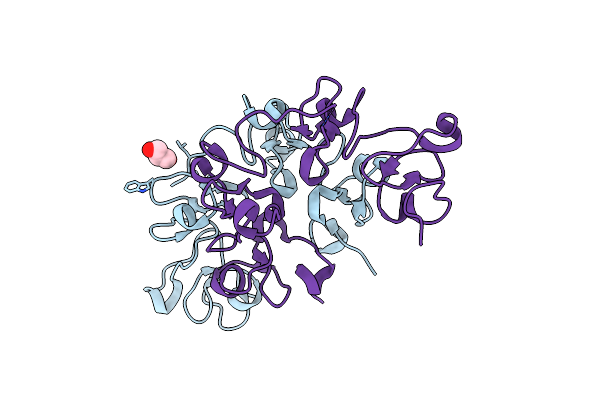
Human Holo Aromatic L-Amino Acid Decarboxylase (Aadc) External Aldimine With L-Dopa Methylester
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-08-30 Classification: LYASE Ligands: PLP, W5I, PG4 |
 |
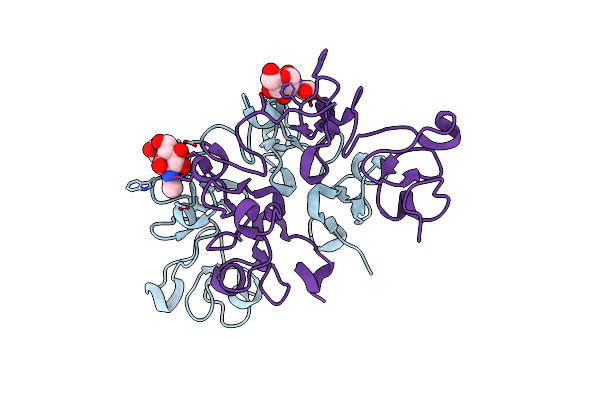
Organism: Arundo donax
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-07-14 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
 |
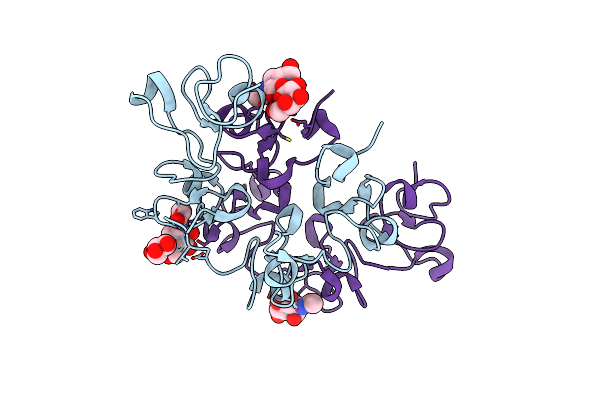
Three Dimensional Structure Of The Giant Reed (Arundodonax) Lectin (Adl) Complex With N-Acetyl Glucosamine
Organism: Arundo donax
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-07-14 Classification: SUGAR BINDING PROTEIN Ligands: NAG, GOL |
 |
Three Dimensional Structure Of The Giant Reed (Arundodonax) Lectin (Adl) Complex With N-Acetyl Lactosamine
Organism: Arundo donax
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-07-14 Classification: SUGAR BINDING PROTEIN Ligands: NAG, GOL |
 |
Three Dimensional Structure Of The Giant Reed (Arundodonax) Lectin (Adl) Complex With Sialic Acid
Organism: Arundo donax
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-07-14 Classification: SUGAR BINDING PROTEIN Ligands: SIA, GOL |
 |
Three Dimensional Structure Of The Giant Reed (Arundodonax) Lectin (Adl) Complex With N,N'-Diacetylchitobiose; 30 Seconds Soaking
Organism: Arundo donax
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-07-14 Classification: SUGAR BINDING PROTEIN Ligands: NAG, GOL |
 |
Three Dimensional Structure Of The Giant Reed (Arundodonax) Lectin (Adl) Complex With N,N'-Diacetylchitobiose; 60 Seconds Soaking
Organism: Arundo donax
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-07-14 Classification: SUGAR BINDING PROTEIN Ligands: NAG, GOL |
 |
Organism: Pleurotus ostreatus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-02-19 Classification: SUGAR BINDING PROTEIN Ligands: NAG, CA, GOL |
 |
Organism: Pleurotus ostreatus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-02-19 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-03-14 Classification: TRANSPORT PROTEIN Ligands: DAO, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-03-07 Classification: TRANSPORT PROTEIN Ligands: PLM, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-03-07 Classification: TRANSPORT PROTEIN Ligands: PLM, CL |

