Search Count: 25
 |
Escherichia Coli D-2-Deoxyribose-5-Phosphate Aldolase - C47V/G204A/S239D Mutant
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2020-11-18 Classification: LYASE Ligands: EDO, FMT, MG |
 |
Escherichia Coli D-2-Deoxyribose-5-Phosphate Aldolase - N21K Mutant Complex With Reaction Products
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2020-11-18 Classification: LYASE Ligands: EDO, MG, G3H |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-11-18 Classification: LYASE Ligands: MG |
 |
Organism: Rhizobium leguminosarum bv. trifolii
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-06-21 Classification: LYASE |
 |
Organism: Rhizobium leguminosarum bv. trifolii (strain wsm2304), Rhizobium leguminosarum bv. trifolii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-06-21 Classification: LYASE Ligands: MG, FES |
 |
Organism: Rhizobium leguminosarum bv. trifolii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-06-21 Classification: LYASE Ligands: FES, MG |
 |
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-03-23 Classification: LYASE Ligands: GOL, FMT |
 |
Crystal Structure Of Keto-Deoxy-D-Galactarate Dehydratase Complexed With 2-Oxoadipic Acid
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-03-23 Classification: LYASE Ligands: OOG, FMT |
 |
Crystal Structure Of Keto-Deoxy-D-Galactarate Dehydratase Complexed With Pyruvate
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-03-23 Classification: LYASE Ligands: PYR, GOL, FMT |
 |
Crystal Structure Of Aldose-Aldose Oxidoreductase From Caulobacter Crescentus Complexed With Glycerol
Organism: Caulobacter crescentus cb15
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-10-21 Classification: OXIDOREDUCTASE Ligands: NAP, GOL, SO4 |
 |
Crystal Structure Of Aldose-Aldose Oxidoreductase From Caulobacter Crescentus Complexed With Xylose
Organism: Caulobacter crescentus cb15
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-10-21 Classification: OXIDOREDUCTASE Ligands: NDP, XYP, SO4, XYS, DIO |
 |
Crystal Structure Of Aldose-Aldose Oxidoreductase From Caulobacter Crescentus Complexed With Glucose
Organism: Caulobacter crescentus cb15
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-10-21 Classification: OXIDOREDUCTASE Ligands: NDP, BGC, DIO, SO4 |
 |
Crystal Structure Of Aldose-Aldose Oxidoreductase From Caulobacter Crescentus Complexed With Maltotriose
Organism: Caulobacter crescentus cb15
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-10-21 Classification: OXIDOREDUCTASE Ligands: NDP, MLI |
 |
Crystal Structure Of Aldose-Aldose Oxidoreductase From Caulobacter Crescentus Complexed With Sorbitol
Organism: Caulobacter crescentus cb15
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2015-10-21 Classification: OXIDOREDUCTASE Ligands: NDP, SOR, SO4 |
 |
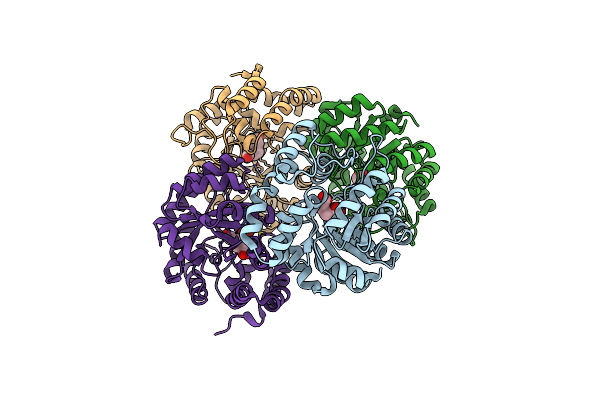
Crystal Structure Of Keto-Deoxy-D-Galactarate Dehydratase Complexed With Pyruvate
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-12-17 Classification: LYASE Ligands: GOL, FMT |
 |
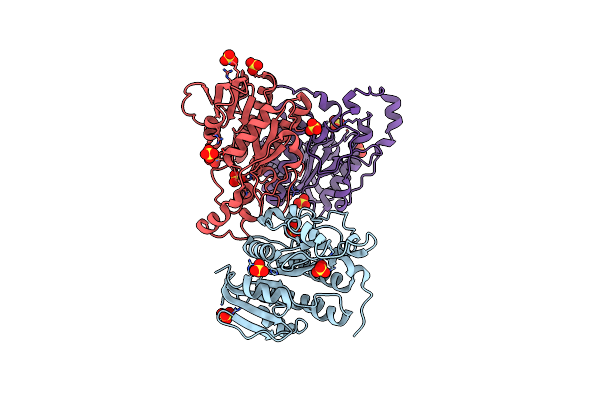
Crystal Structure Of Keto-Deoxy-D-Galactarate Dehydratase Complexed With 2-Oxoadipic Acid
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-12-17 Classification: LYASE Ligands: OOG, FMT |
 |
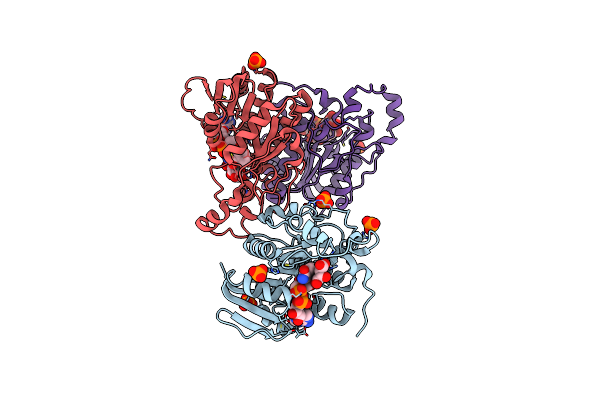
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-06-15 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
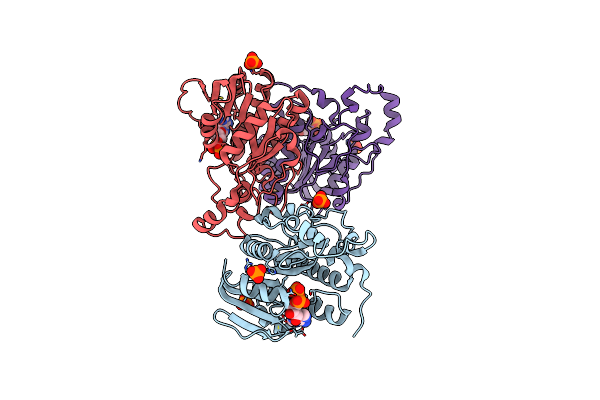
Crystal Structure Of Uronate Dehydrogenase From Agrobacterium Tumefaciens Complexed With Nadh And Product
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-06-15 Classification: OXIDOREDUCTASE Ligands: NAI, 15L, PO4 |
 |
Crystal Structure Of Uronate Dehydrogenase From Agrobacterium Tumefaciens, Y136A Mutant Complexed With Nad
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-06-15 Classification: OXIDOREDUCTASE Ligands: NAD, PO4 |
 |
Atomic Resolution Structure Of The Hfbii Hydrophobin: A Self-Assembling Amphiphile
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2004-01-13 Classification: surface active protein Ligands: MN |

