Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
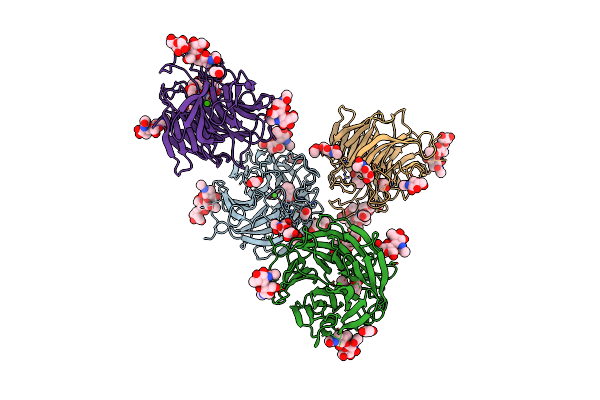 |
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-04-05 Classification: BIOSYNTHETIC PROTEIN Ligands: NAG, CA, BEZ, EDO, PEG, GOL, MAN |
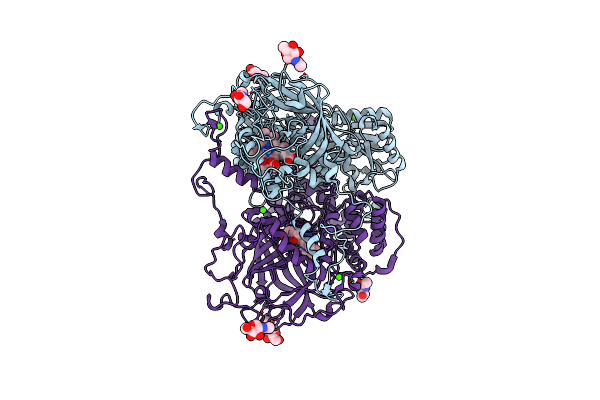 |
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-06-16 Classification: OXIDOREDUCTASE Ligands: HDD, NAG, 3TR, CA |
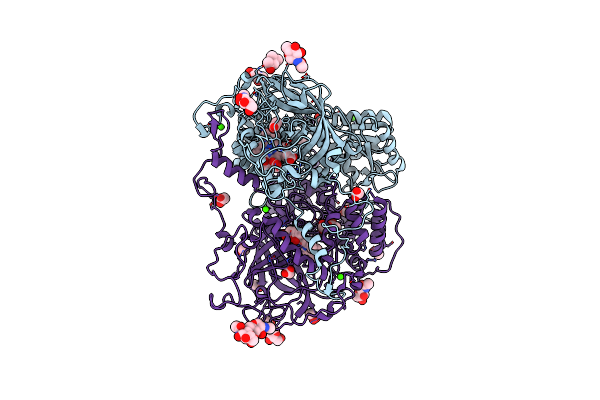 |
The Structures Of Penicillium Vitale Catalase: Resting State, Oxidised State (Compound I) And Complex With Aminotriazole
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2006-07-10 Classification: OXIDOREDUCTASE Ligands: HDD, NAG, O, CA, ACT, F50, MPD |
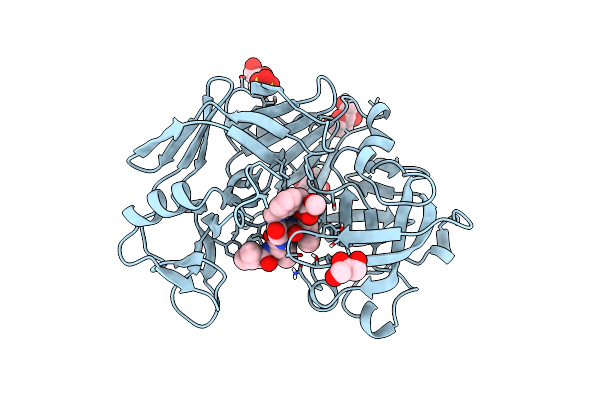 |
Acid Proteinase (Penicillopepsin) (E.C.3.4.23.20) Complex With Phosphonate Inhibitor: Methyl Cyclo[(2S)-2-[[(1R)-1-(N-(L-N-(3-Methylbutanoyl)Valyl-L-Aspartyl)Amino)-3-Methylbut Yl] Hydroxyphosphinyloxy]-3-(3-Aminomethyl) Phenylpropanoate
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 1998-10-14 Classification: HYDROLASE Ligands: MAN, SO4, PP7, GOL |
 |
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 1998-10-14 Classification: HYDROLASE Ligands: MAN, SO4, ACT, PP8, GOL |
 |
Acid Proteinase (Penicillopepsin) (E.C.3.4.23.20) Complex With Phosphonate Inhibitor: Methyl[Cyclo-7[(2R)-((N-Valyl) Amino)-2-(Hydroxyl-(1S)-1-Methyoxycarbonyl-2-Phenylethoxy) Phosphinyloxy-Ethyl]-1-Naphthaleneacetamide], Sodium Salt
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 1998-05-27 Classification: HYDROLASE Ligands: MAN, SO4, PP6 |
 |
Acid Proteinase (Penicillopepsin) (E.C.3.4.23.20) Complex With Phosphonate Inhibitor: Methyl(2S)-[1-(((N-Formyl)-L-Valyl)Amino-2-(2-Naphthyl)Ethyl)Hydroxyphosphinyloxy]-3-Phenylpropanoate, Sodium Salt
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 1998-05-27 Classification: HYDROLASE Ligands: MAN, SO4, PP4 |
 |
Acid Proteinase (Penicillopepsin) (E.C.3.4.23.20) Complex With Phosphonate Inhibitor: Methyl(2S)-[1-(((N-(1-Naphthaleneacetyl))-L-Valyl)Aminomethyl)Hydroxy Phosphinyloxy]-3-Phenylpropanoate, Sodium Salt
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 1998-05-27 Classification: HYDROLASE Ligands: MAN, SO4, PP5 |
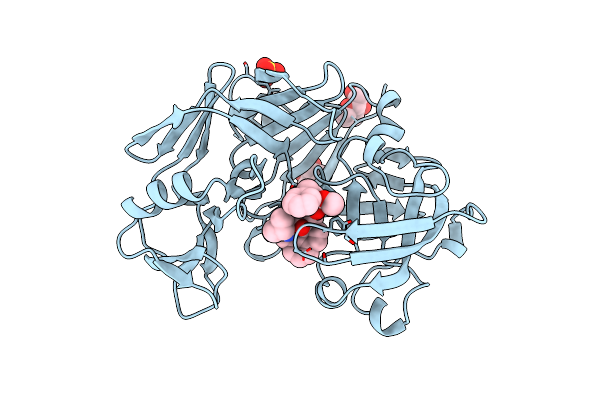 |
Acid Proteinase (Penicillopepsin) (E.C.3.4.23.20) Complex With Phosphonate Macrocyclic Inhibitor:Methyl[Cyclo-7[(2R)-((N-Valyl)Amino)-2-(Hydroxyl-(1S)-1-Methyoxycarbonyl-2-Phenylethoxy)Phosphinyloxy-Ethyl]-1-Naphthaleneacetamide], Sodium Salt
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 1998-05-27 Classification: HYDROLASE Ligands: MAN, SO4, PP6 |
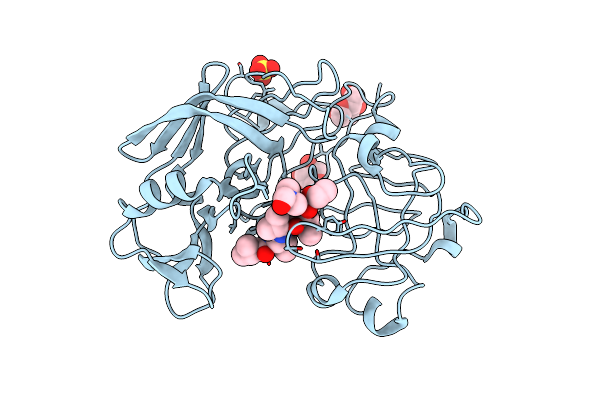 |
Crystallographic Analysis Of Transition State Mimics Bound To Penicillopepsin: Phosphorous-Containing Peptide Analogues
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-05-31 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: IVV, MAN, HSY, SO4, DMF |
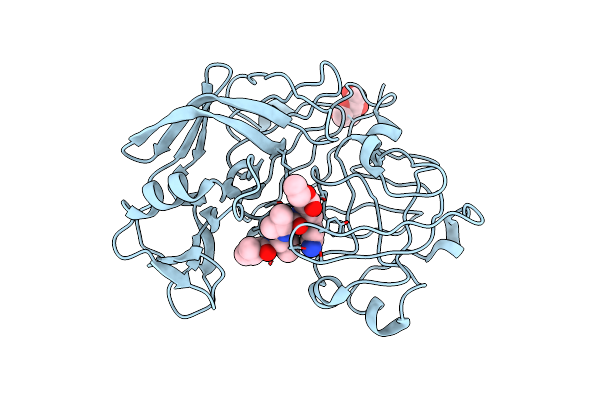 |
Crystallographic Analysis Of A Pepstatin Analogue Binding To The Aspartyl Proteinase Penicillopepsin At 1.8 Angstroms Resolution
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-01-31 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MAN |
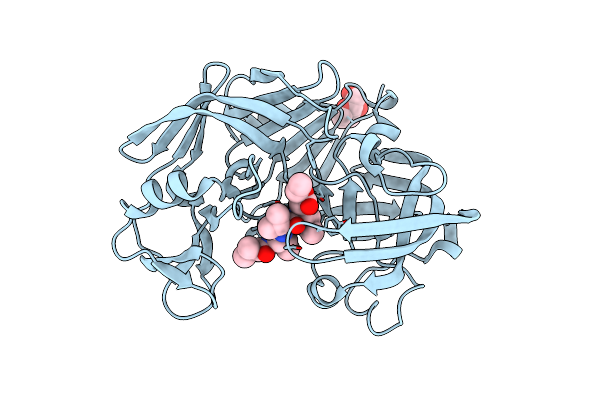 |
Crystallographic Analysis Of A Pepstatin Analogue Binding To The Aspartyl Proteinase Penicillopepsin At 1.8 Angstroms Resolution
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-01-31 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MAN |
 |
Crystallographic Analysis Of Transition State Mimics Bound To Penicillopepsin: Difluorostatine-And Difluorostatone-Containing Peptides
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-01-31 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MAN, XYS, SO4, DMF |
 |
Crystallographic Analysis Of Transition State Mimics Bound To Penicillopepsin: Difluorostatine-And Difluorostatone-Containing Peptides
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-01-31 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MAN, HSY, SO4, DMF |
 |
Crystal Structure Of The Fungal Peroxidase From Arthromyces Ramosus At 1.9 Angstroms Resolution: Structural Comparisons With The Lignin And Cytochrome C Peroxidases
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1994-01-31 Classification: PEROXIDASE(DONOR:H2O2 OXIDOREDUCTASE) Ligands: CA, HEM |
 |
Crystallographic Analysis Of Transition-State Mimics Bound To Penicillopepsin: Phosphorus-Containing Peptide Analogues
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1993-10-31 Classification: HYDROLASE/hydrolase inhibitor Ligands: 1Z7, MAN, XYS, SO4 |
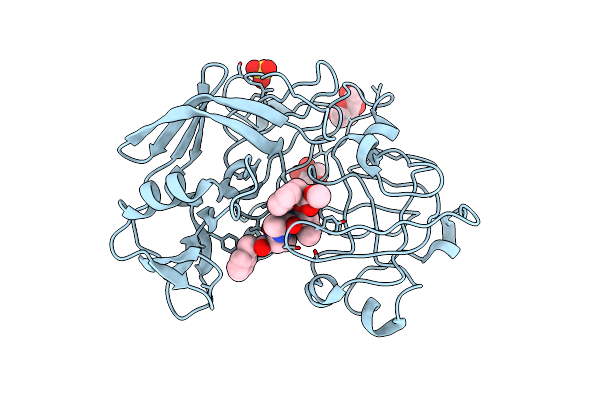 |
Crystallographic Analysis Of Transition-State Mimics Bound To Penicillopepsin: Phosphorus-Containing Peptide Analogues
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1993-10-31 Classification: HYDROLASE/hydrolase inhibitor Ligands: 0P1, MAN, ARA, SO4 |
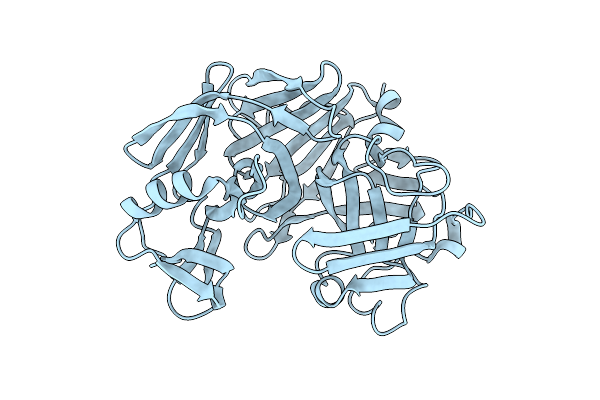 |
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1991-01-15 Classification: HYDROLASE (ACID PROTEINASE) |
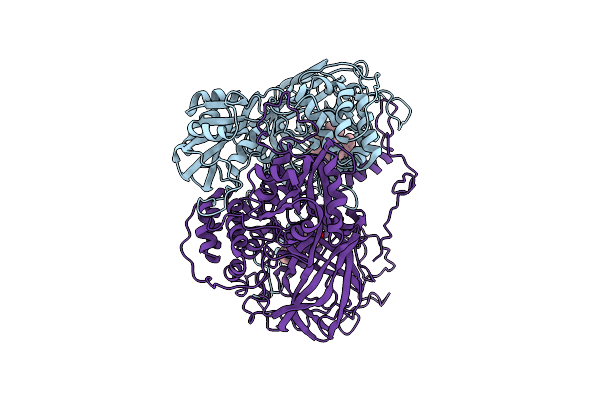 |
Three-Dimensional Structure Of Catalase From Penicillium Vitale At 2.0 Angstroms Resolution
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1983-09-06 Classification: OXIDOREDUCTASE(H2O2(A)) Ligands: HEM |
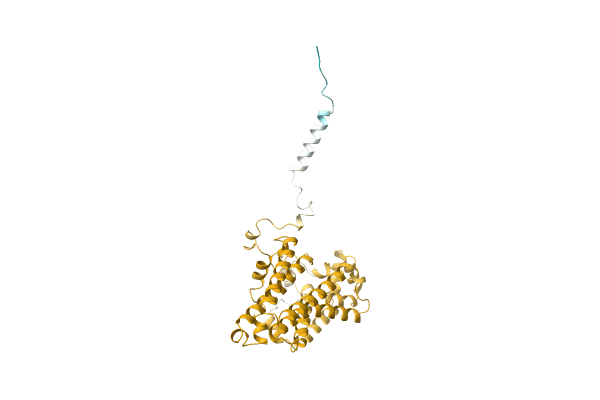 |