Search Count: 182
 |
Conformational Flexibility In Hla-B8: Peptide Tuning Structural And Dynamical Changes
Organism: Homo sapiens, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Conformational Flexibility In Hla-B8: Peptide Tuning Structural And Dynamical Changes
Organism: Homo sapiens, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Conformational Flexibility In Hla-B8: Peptide Tuning Structural And Dynamical Changes
Organism: Homo sapiens, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
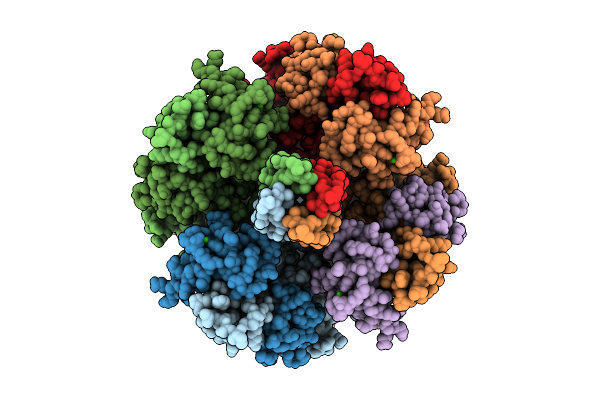 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PT5, K, CA |
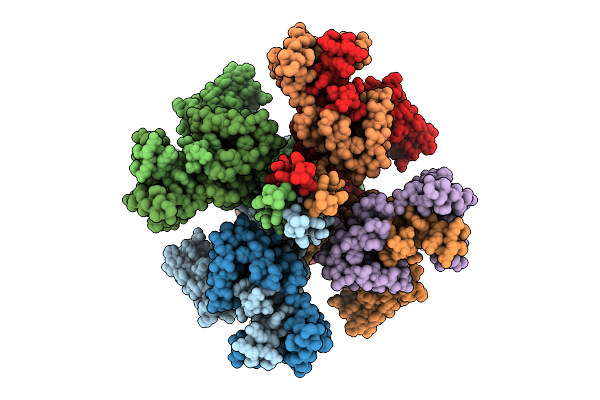 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PIO, K |
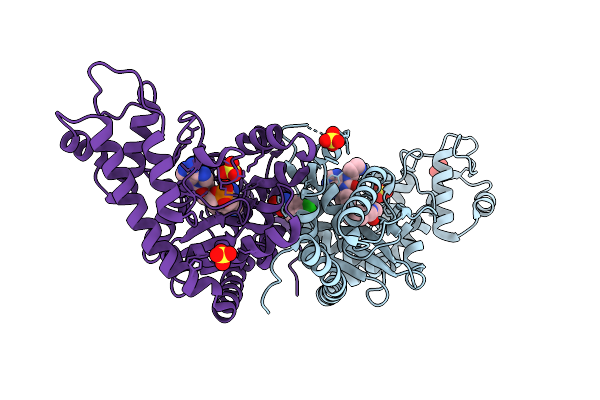 |
Crystal Structure Of Escherichia Coli Tryptophanyl-Trna Synthetase In Complex With 71G
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2025-09-24 Classification: LIGASE Ligands: A1ERS, SO4, TYM, CLW |
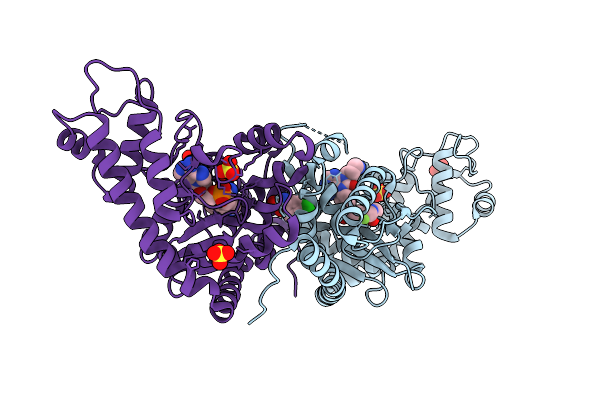 |
Crystal Structure Of Escherichia Coli Tryptophanyl-Trna Synthetase In Complex With An Inhibitor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-09-24 Classification: LYASE Ligands: A1ERR, SO4, CLW, TYM |
 |
Crystal Structure Of Escherichia Coli Trptophanyl-Trna Synthetase In Complex With N-Piperidine Ibrutinib
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2025-05-07 Classification: LIGASE Ligands: A1EKY, SO4, CLW, TYM |
 |
Crystal Structure Of Escherichia Coli Trptophanyl-Trna Synthetase In Complex With Trna(Trp)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2025-05-07 Classification: LIGASE/RNA |
 |
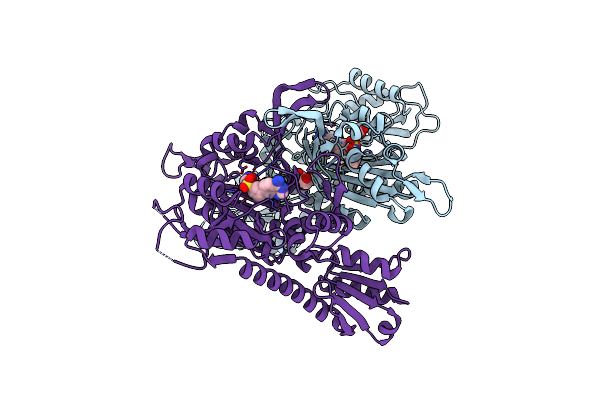
Crystal Structure Of Escherichia Coli Trptophanyl-Trna Synthetase In Complex With Tirabrutinib
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-05-07 Classification: LIGASE Ligands: A1EK0, SO4, CLW, TYM |
 |
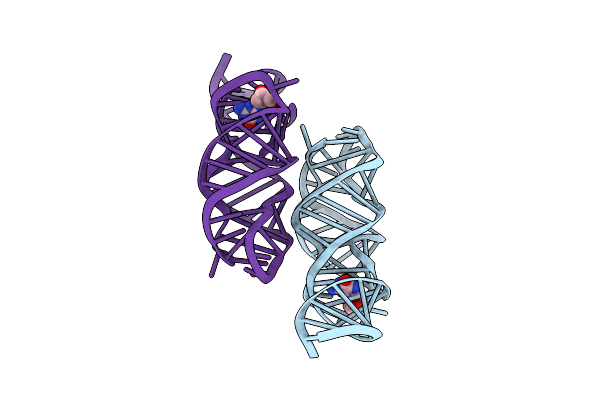
Crystal Structure Of Human Prolyl-Trna Synthetase In Complex With Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-04-02 Classification: LIGASE Ligands: W2H, EDO |
 |
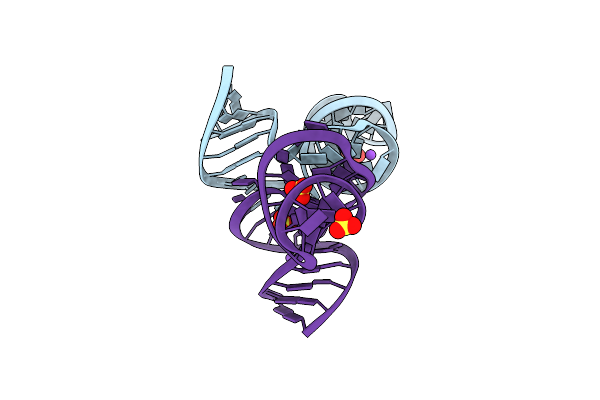
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2025-03-19 Classification: RNA Ligands: GNG |
 |
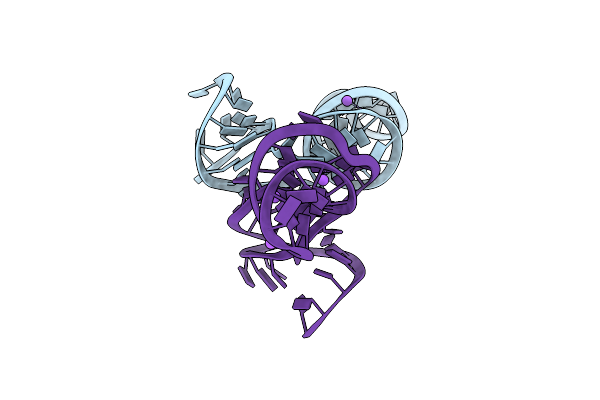
Organism: Homo
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2025-03-19 Classification: RNA Ligands: SO4, NA |
 |
Organism: Homo
Method: X-RAY DIFFRACTION Resolution:3.86 Å Release Date: 2025-03-19 Classification: RNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-03-19 Classification: RNA Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2025-03-19 Classification: RNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-03-19 Classification: RNA Ligands: GUN, BA, PO4, NA |
 |
Crystal Structure Of The Mbp-Mcl1 Complex With Highly Selective And Potent Cyclic Peptide Inhibitor
Organism: Escherichia coli k-12, Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2025-03-19 Classification: APOPTOSIS |
 |
Organism: Enterococcus faecalis
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RNA |
 |
Organism: Enterococcus faecalis jh1
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RNA |

