Search Count: 21
 |
Organism: Streptomyces clavuligerus (strain atcc 27064 / dsm 738 / jcm 4710 / nbrc 13307 / ncimb 12785 / nrrl 3585 / vkm ac-602)
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2018-10-31 Classification: LYASE |
 |
Organism: Streptomyces clavuligerus (strain atcc 27064 / dsm 738 / jcm 4710 / nbrc 13307 / ncimb 12785 / nrrl 3585 / vkm ac-602)
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2018-10-31 Classification: LYASE |
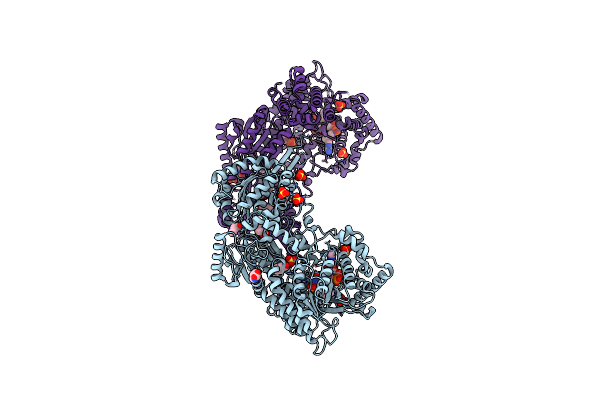 |
Structure Of Proline Utilization A (Puta) With Proline Bound In Remote Sites
Organism: Bradyrhizobium diazoefficiens (strain jcm 10833 / iam 13628 / nbrc 14792 / usda 110)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-01-03 Classification: OXIDOREDUCTASE Ligands: FDA, PRO, SO4 |
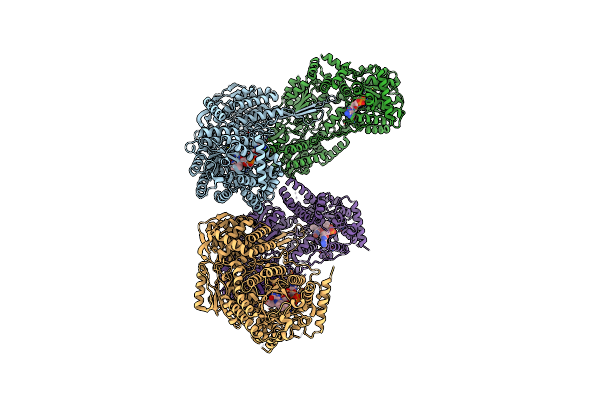 |
Crystal Structure Of Proline Utilization A (Puta) From Bdellovibrio Bacteriovorus Inactivated By N-Propargylglycine
Organism: Bdellovibrio bacteriovorus (strain atcc 15356 / dsm 50701 / ncib 9529 / hd100)
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2017-08-02 Classification: OXIDOREDUCTASE Ligands: P5F |
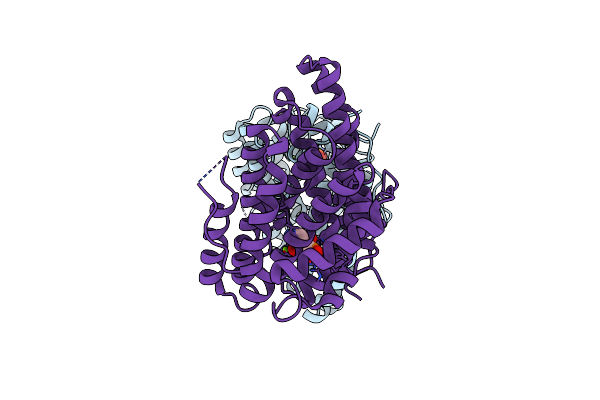 |
Geosmin Synthase From Streptomyces Coelicolor N-Terminal Domain Complexed With Three Mg2+ Ions And Alendronic Acid
Organism: Streptomyces coelicolor (strain atcc baa-471 / a3(2) / m145)
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2015-12-09 Classification: LYASE Ligands: MG, 212 |
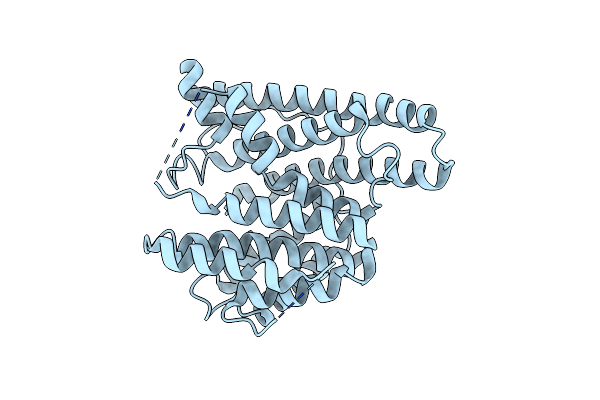 |
Crystal Structure Of The Unliganded Geosmin Synthase N-Terminal Domain From Streptomyces Coelicolor
Organism: Streptomyces coelicolor (strain atcc baa-471 / a3(2) / m145)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-11-25 Classification: LYASE |
 |
Crystal Structure Of Bradyrhizobium Japonicum Proline Utilization A (Puta) Mutant D779W
Organism: Bradyrhizobium diazoefficiens usda 110
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-08-06 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, GOL |
 |
Crystal Structure Of Bradyrhizobium Japonicum Proline Utilization A (Puta) Mutant D779Y
Organism: Bradyrhizobium diazoefficiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-08-06 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, SO4 |
 |
Crystal Structure Of Bradyrhizobium Japonicum Proline Utilization A (Puta) Mutant D778Y
Organism: Bradyrhizobium diazoefficiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-08-06 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, GOL |
 |
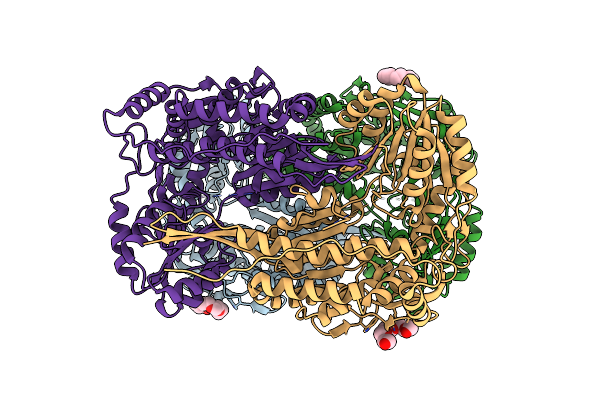
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2014-02-19 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
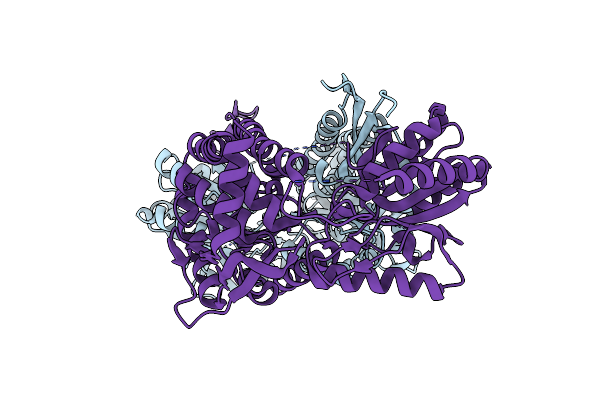
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-02-19 Classification: OXIDOREDUCTASE Ligands: 1PE, MG |
 |
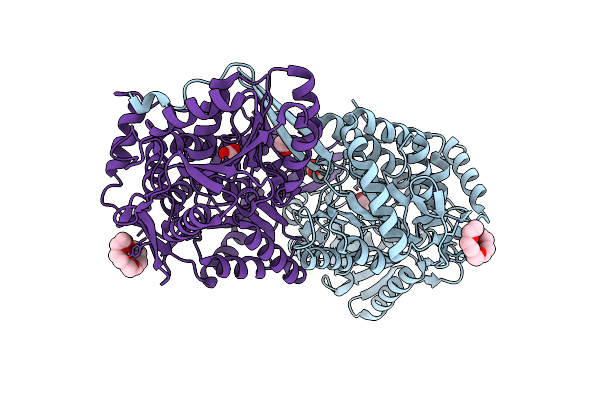
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-02-19 Classification: OXIDOREDUCTASE |
 |
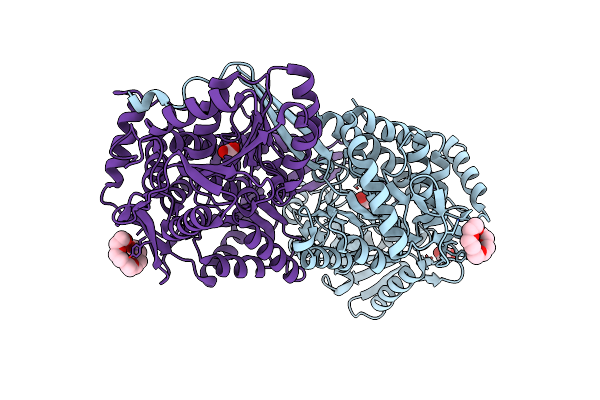
Structure Of Mouse 1-Pyrroline-5-Carboxylate Dehydrogenase (Aldh4A1) Complexed With Acetate
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: ACY, 1PE |
 |
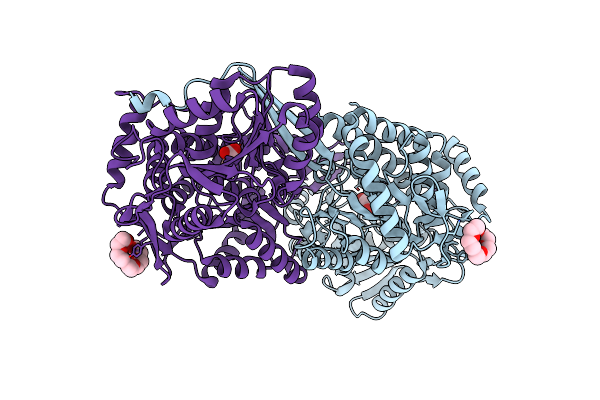
Structure Of Mouse 1-Pyrroline-5-Carboxylate Dehydrogenase (Aldh4A1) Complexed With Glyoxylate
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: GLV, 1PE |
 |
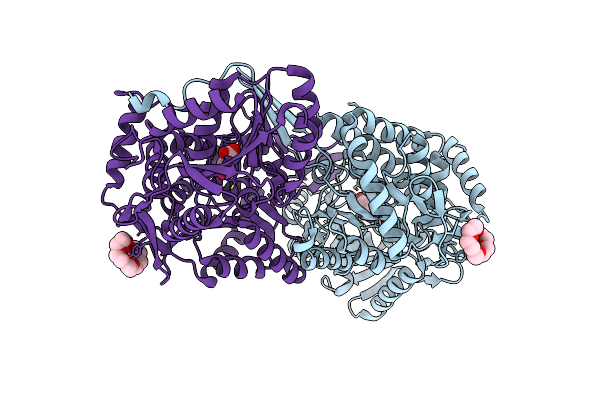
Structure Of Mouse 1-Pyrroline-5-Carboxylate Dehydrogenase (Aldh4A1) Complexed With Malonate
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: MLA, 1PE |
 |
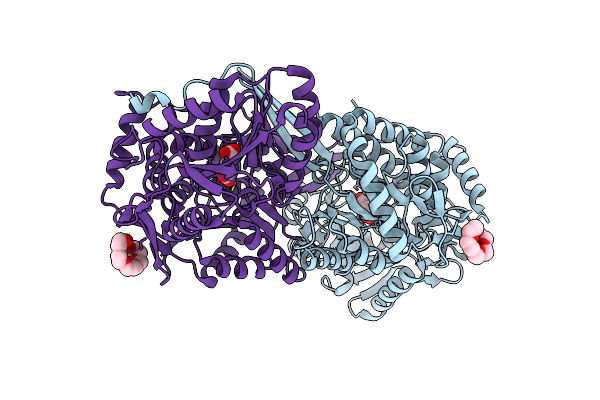
Structure Of Mouse 1-Pyrroline-5-Carboxylate Dehydrogenase (Aldh4A1) Complexed With Succinate
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: 1PE, SIN |
 |
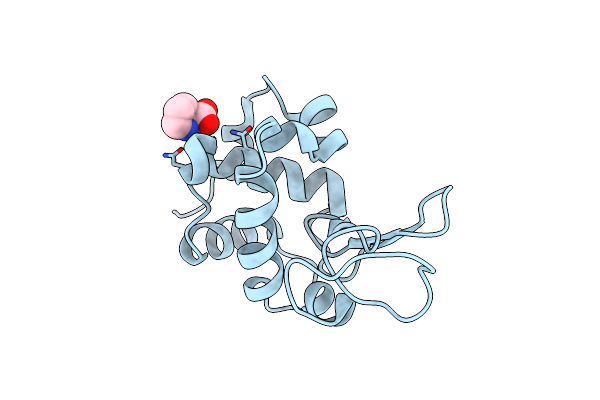
Structure Of Mouse 1-Pyrroline-5-Carboxylate Dehydrogenase (Aldh4A1) Complexed With Glutarate
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2013-08-28 Classification: OXIDOREDUCTASE Ligands: 1PE, GUA |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-07-25 Classification: HYDROLASE Ligands: PRO, CL |
 |
Crystal Structure Of Xylose Isomerase From Streptomyces Rubiginosus Cryoprotected In Proline
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-07-25 Classification: ISOMERASE Ligands: PRO, SO4, MG, MN |
 |
Crystal Structure Francisella Tularensis Histidine Acid Phosphatase Cryoprotected With Proline
Organism: Francisella tularensis subsp. holarctica
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2012-07-25 Classification: HYDROLASE Ligands: SO4, PRO |

