Search Count: 50
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2025-02-19 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2025-02-19 Classification: HYDROLASE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-02-19 Classification: HYDROLASE Ligands: ACY, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-02-19 Classification: HYDROLASE Ligands: ACT |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2025-02-19 Classification: HYDROLASE Ligands: PG4, TLA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-19 Classification: DE NOVO PROTEIN Ligands: PG4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2024-11-20 Classification: DE NOVO PROTEIN Ligands: HCY, SO4 |
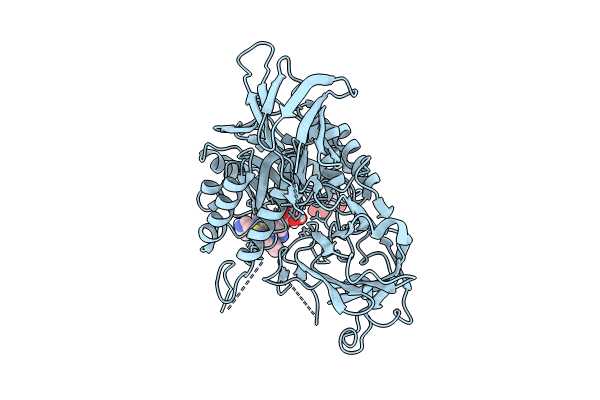 |
Organism: Lachnospira eligens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-07-24 Classification: HYDROLASE Ligands: WOF, WR8, GOL |
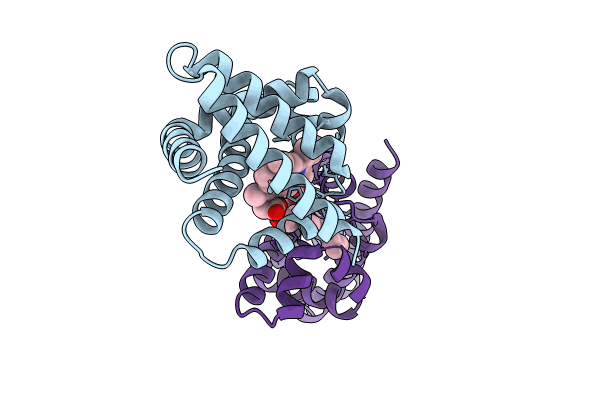 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: DE NOVO PROTEIN Ligands: HEM |
 |
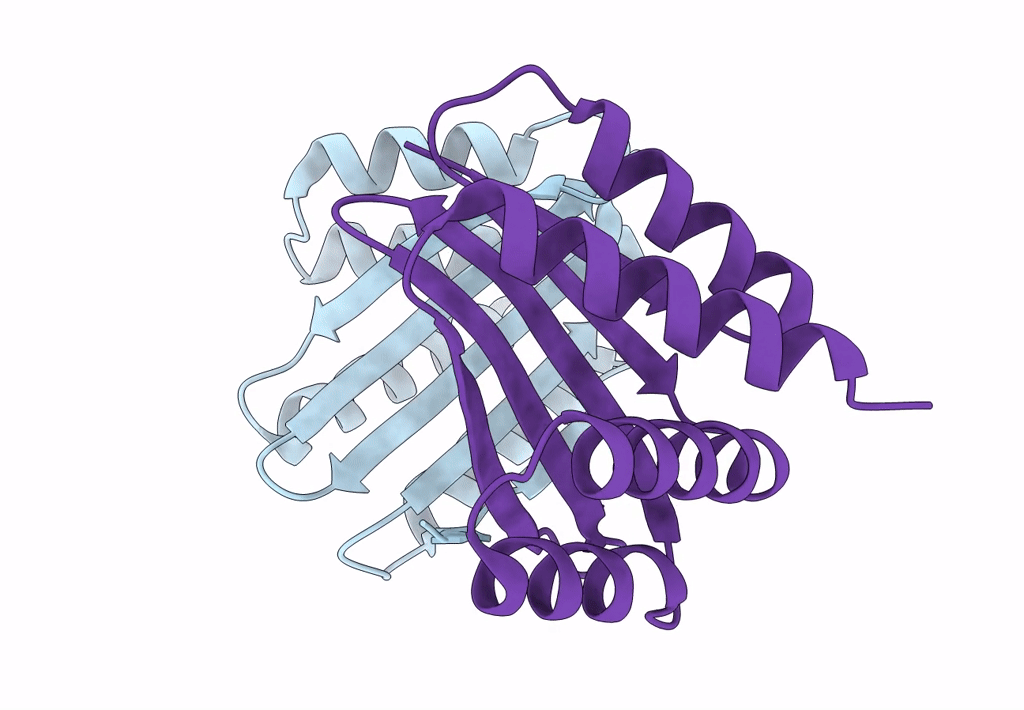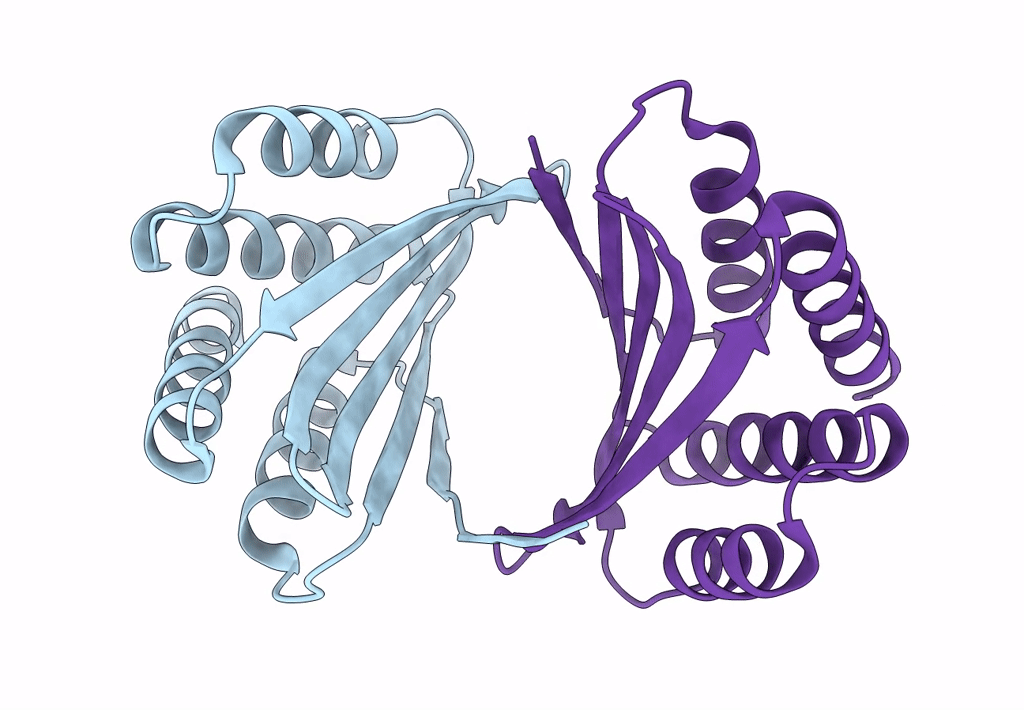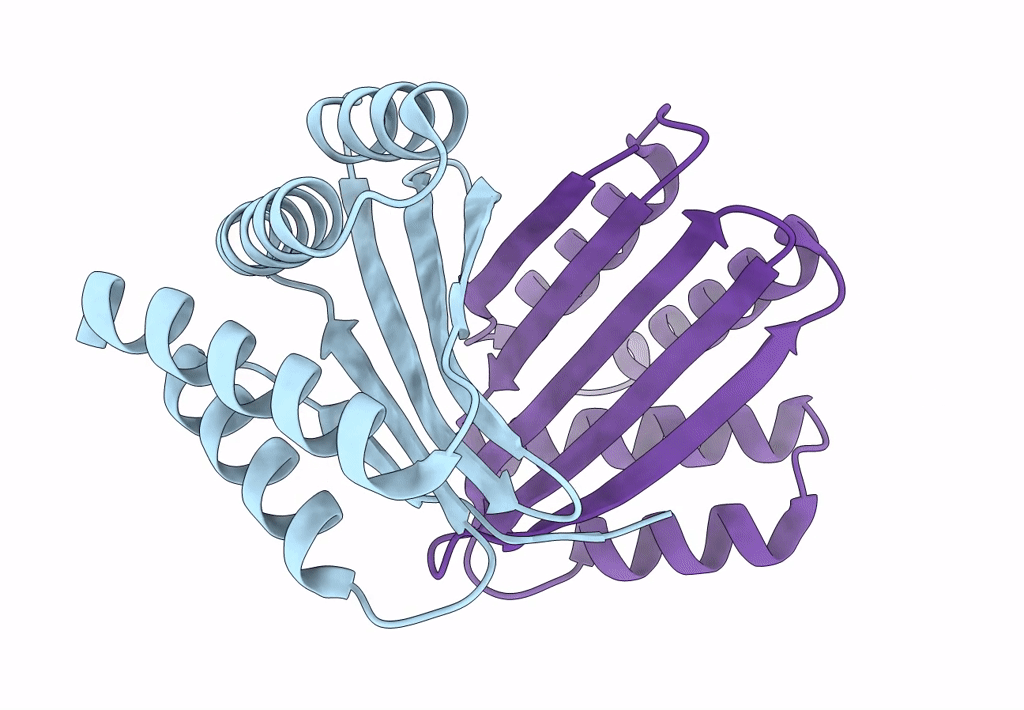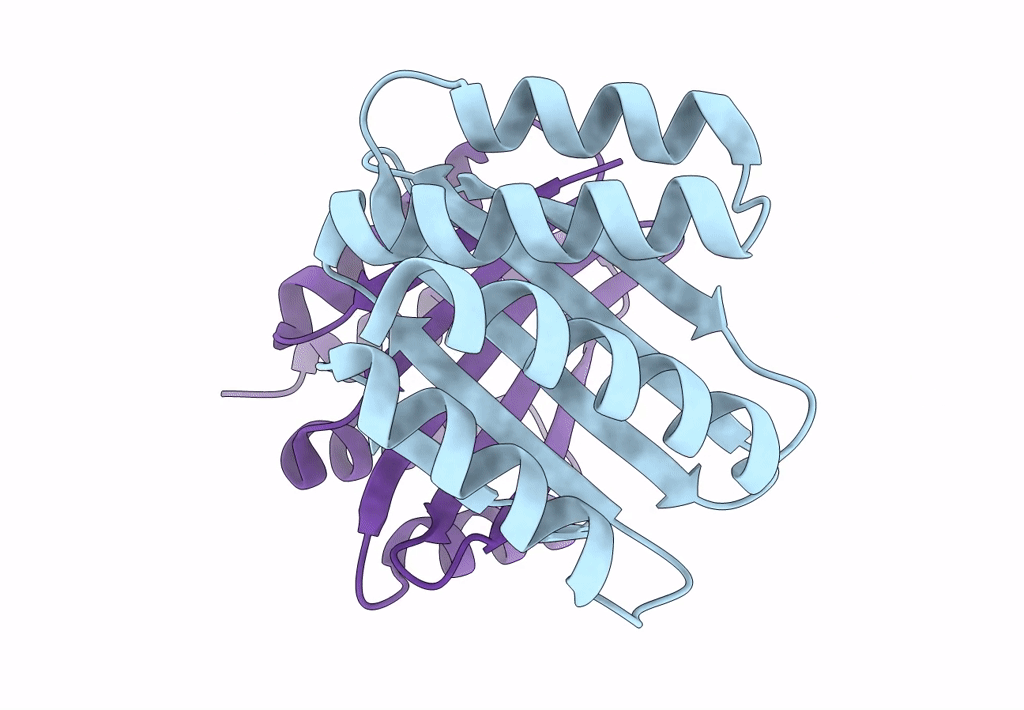
Cryo-Em Structure Of Designed Influenza Ha Binder, Ha_20, Bound To Influenza Ha (Strain: Iowa43)
Organism: Influenza a virus, Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: DE NOVO PROTEIN/Viral Protein Ligands: NAG |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-09-28 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-12-29 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-12-22 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2021-12-15 Classification: DE NOVO PROTEIN |
 |
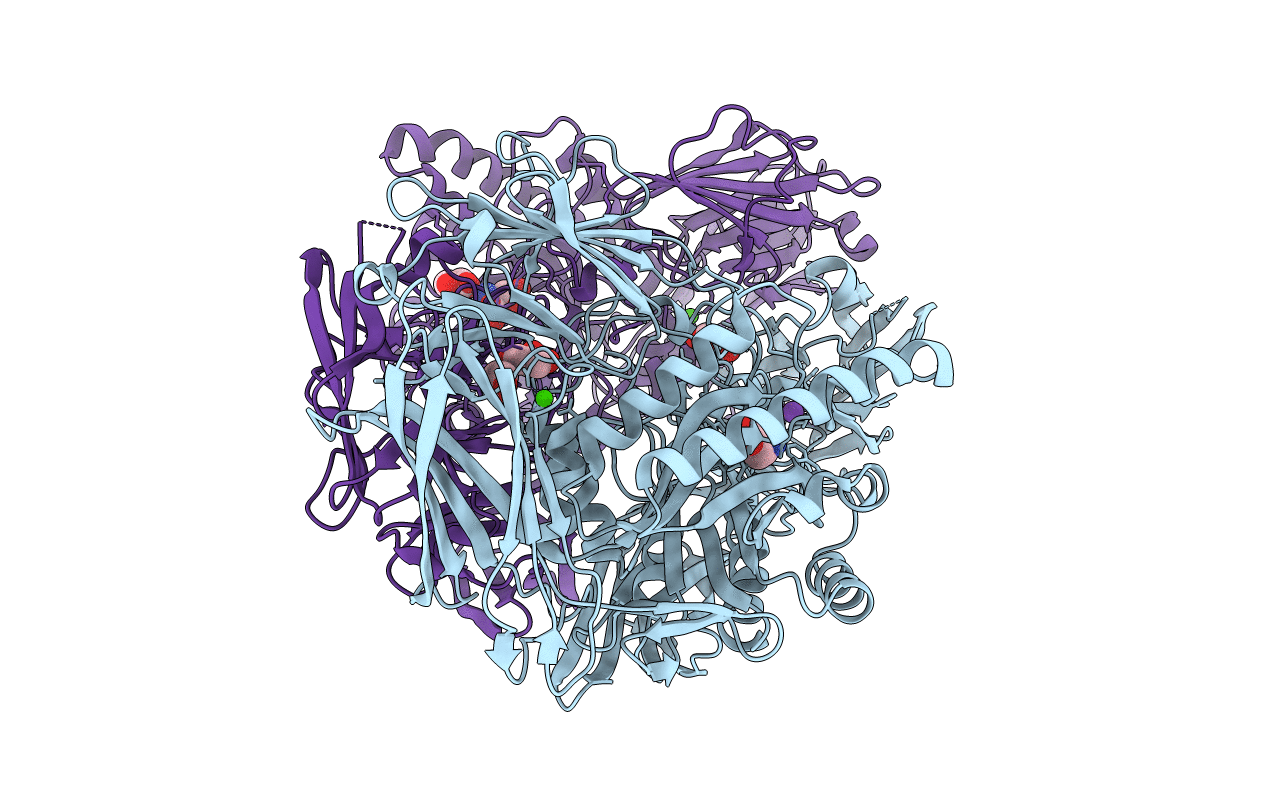
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-04-01 Classification: TRANSCRIPTION Ligands: NQD, DMS |
 |
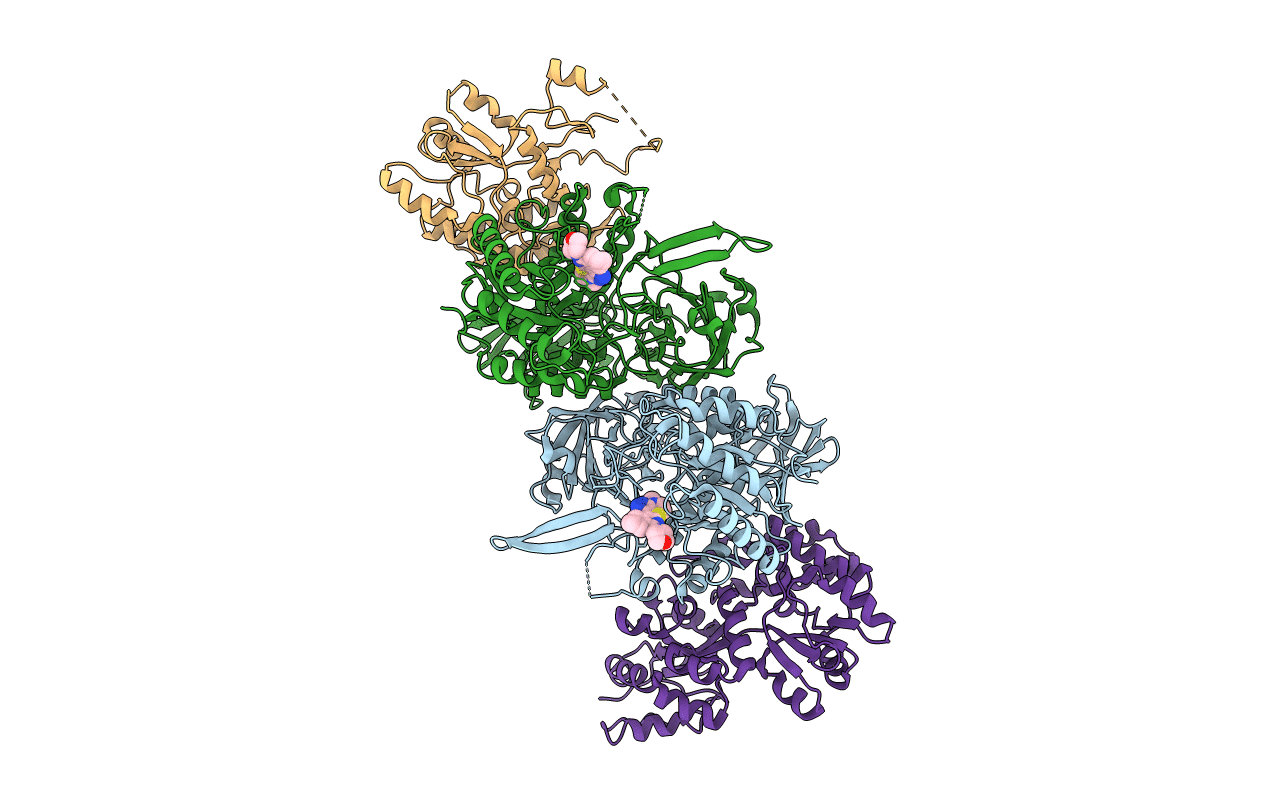
Bacteroides Uniformis Beta-Glucuronidase 2 Covalently Bound To Cyclophellitol-6-Carboxylate Aziridine
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2019-12-18 Classification: hydrolase/hydrolase inhibitor Ligands: L9D, GOL, K, CA |
 |
Crystal Structure Of Clostridium Perfringens Beta-Glucuronidase Bound With A Novel, Potent Inhibitor 4-(8-(Piperazin-1-Yl)-1,2,3,4-Tetrahydro-[1,2,3]Triazino[4',5':4,5]Thieno[2,3-C]Isoquinolin-5-Yl)Morpholine
Organism: Clostridium perfringens (strain 13 / type a), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-04-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: FJV |
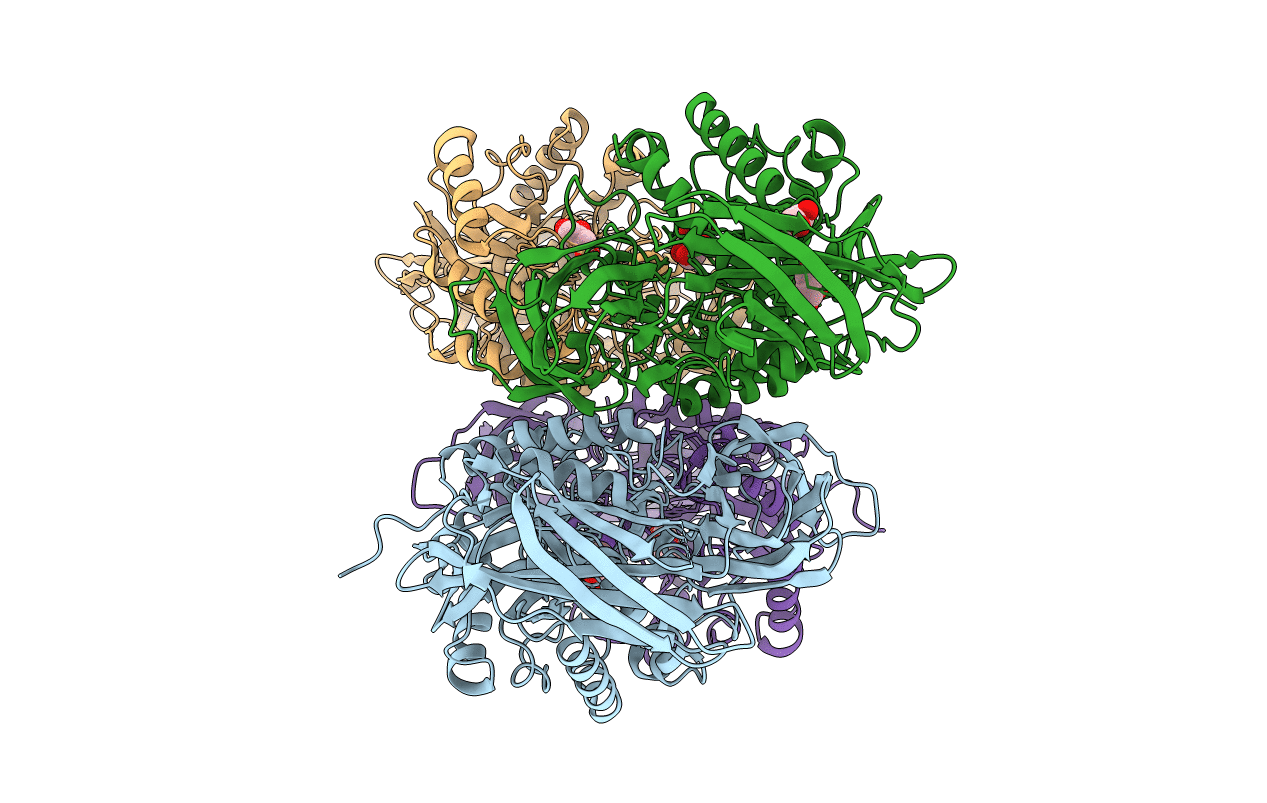 |
Crystal Structure Of A Gh2 Beta-Galacturonidase From Eisenbergiella Tayi Bound To Glycerol
Organism: Eisenbergiella tayi
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-02-20 Classification: HYDROLASE Ligands: CL, GOL |
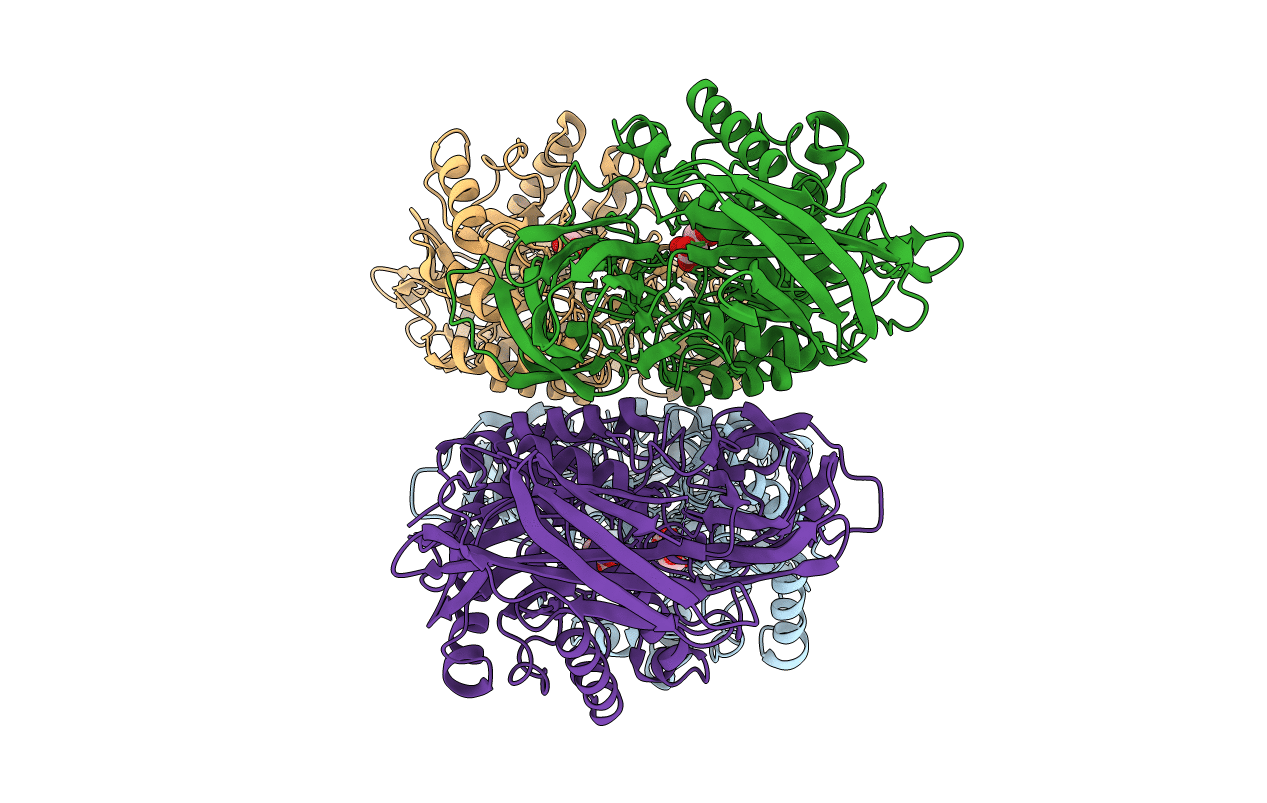 |
Crystal Structure Of Gh2 Beta-Galacturonidase From Eisenbergiella Tayi Bound To Galacturonate
Organism: Eisenbergiella tayi
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-02-20 Classification: HYDROLASE Ligands: ADA, CL |
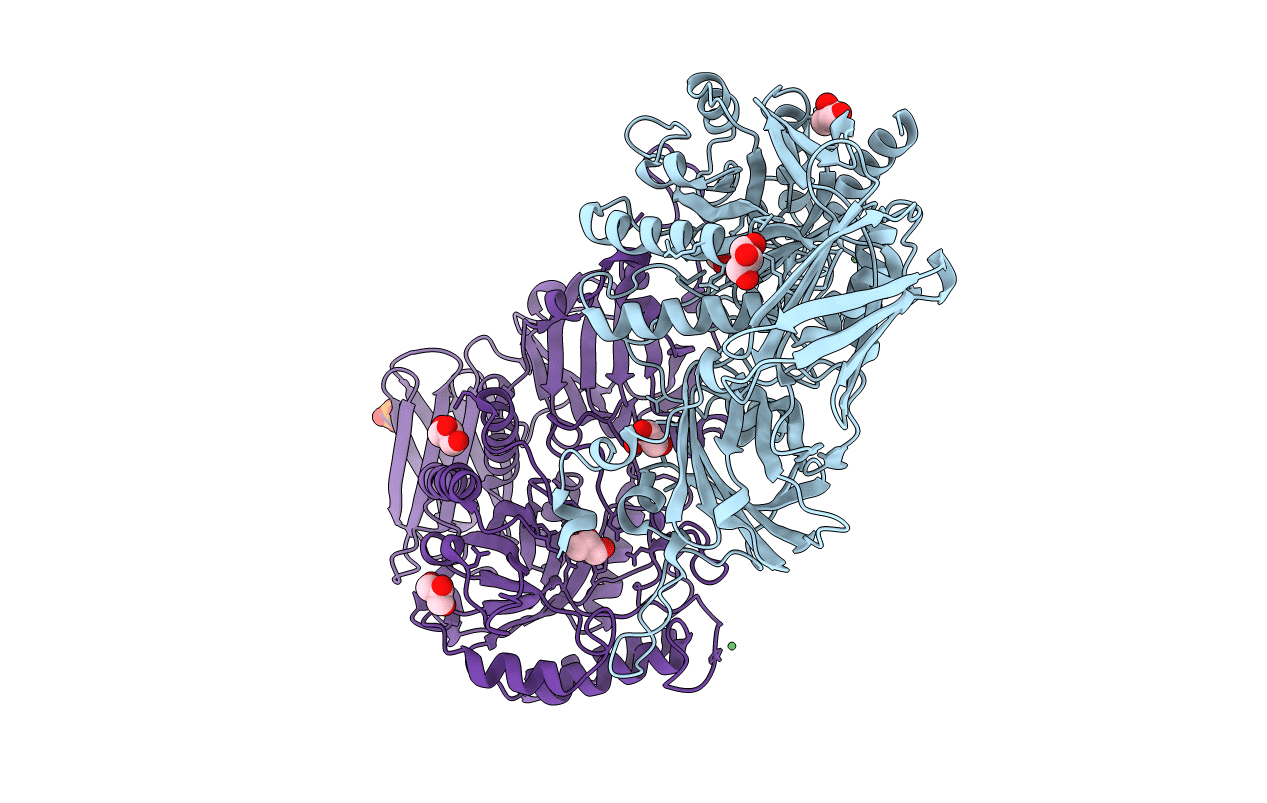 |
Crystal Structure Of Hybrid Beta-Glucuronidase/Beta-Galacturonidase From Fusicatenibacter Saccharivorans
Organism: Fusicatenibacter saccharivorans
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-02-20 Classification: HYDROLASE Ligands: NI, GOL, PO4 |

