Search Count: 32
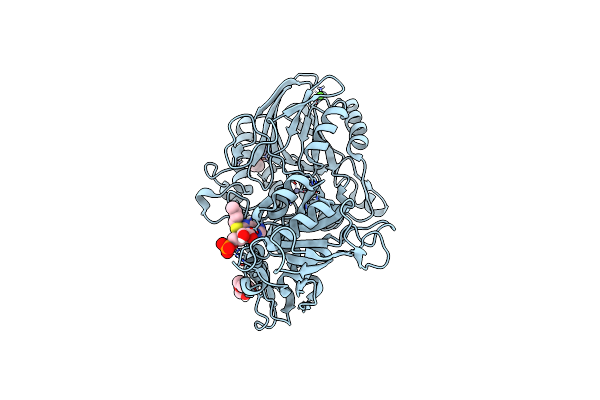 |
Crystal Structure Of Cd73 (Ecto-5'-Nucleotidase) Complexed To 8-Butylthioadenosine 5'Monophosphate (Compound 3 In Publication) In The Open Enzyme State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: ZN, CA, A1IVB, 1PE, PGE |
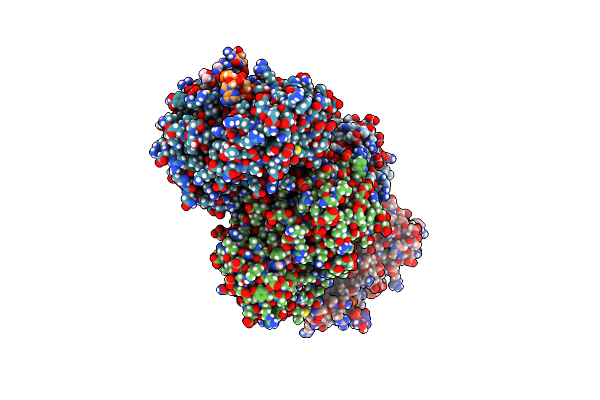 |
Crystal Structure Of The Human Eif4A1/Amppnp/Amidino-Rocaglate/Polypurine Rna Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2025-03-12 Classification: TRANSLATION, HYDROLASE/RNA Ligands: MG, ANP, A1BB1 |
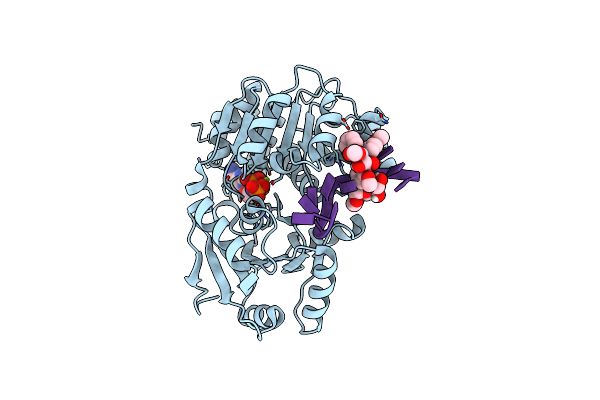 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2024-10-23 Classification: RNA BINDING PROTEIN Ligands: ANP, MG, IOD, A1AG8 |
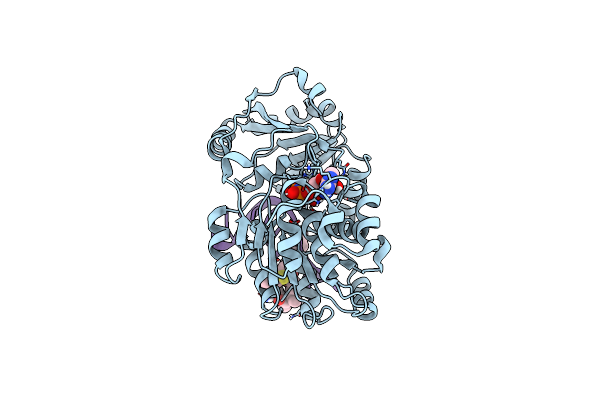 |
Crystal Structure Of Eif4A-I In Complex With Rna Bound To Des-Mepatea, A Pateamine A Analog
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2021-01-06 Classification: TRANSLATION/RNA/INHIBITOR Ligands: ANP, MG, V6D |
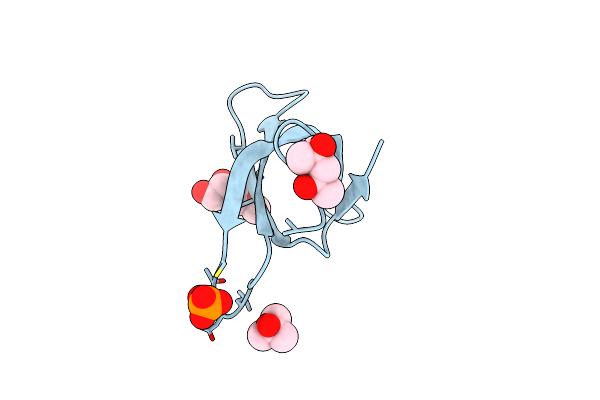 |
Crystal Structure Of Trimethoprim-Resistant Type Ii Dihydrofolate Reductase In Complex With A Bisbenzimidazole Inhibitor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-05-29 Classification: OXIDOREDUCTASE Ligands: D49, MRD, PO4 |
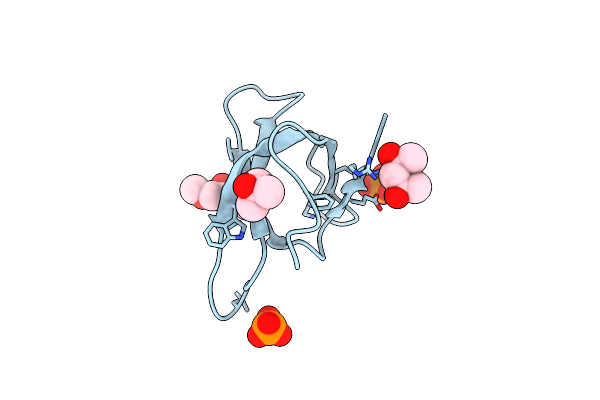 |
Crystal Structure Of Trimethoprim-Resistant Type Ii Dihydrofolate Reductase In Complex With A Bisbenzimidazole Inhibitor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-05-29 Classification: OXIDOREDUCTASE Ligands: LBA, MRD, PO4 |
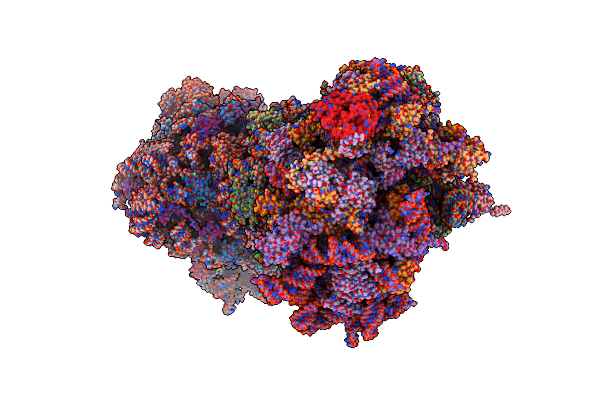 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-07-26 Classification: RIBOSOME Ligands: OHX, MG, 7AL, ZN |
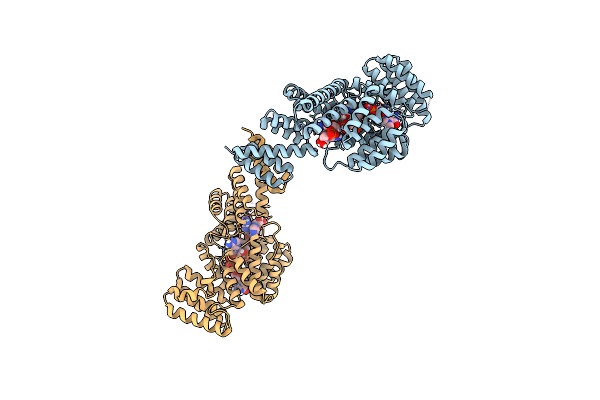 |
Organism: Homo sapiens, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2017-07-19 Classification: RNA binding protein/RNA |
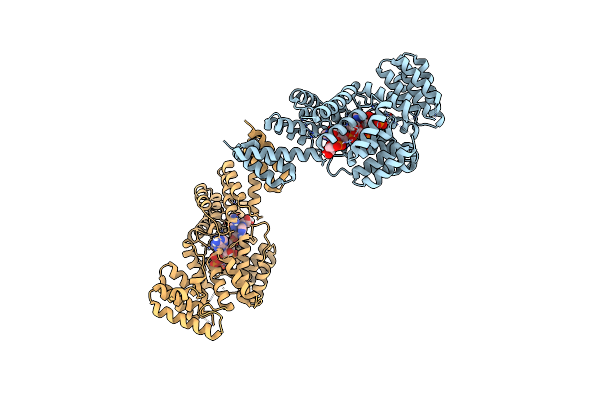 |
Organism: Homo sapiens, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-06-28 Classification: RNA binding protein/RNA |
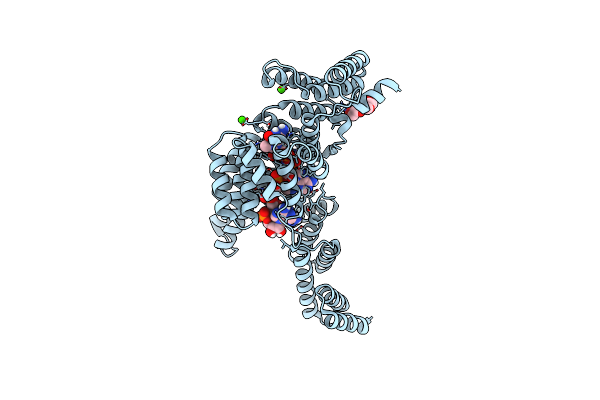 |
Ifit1 Monomeric Mutant (L457E/L464E) With M7Gppp-Aaaa (Syn And Anti Conformations Of Cap)
Organism: Homo sapiens, Unidentified
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2017-03-01 Classification: RNA BINDING PROTEIN Ligands: PGE, CA |
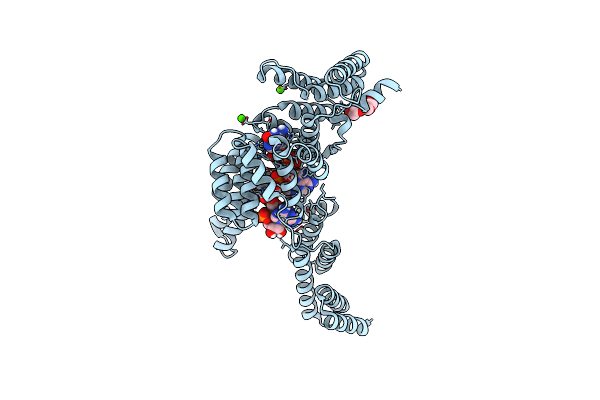 |
Organism: Homo sapiens, Unidentified
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2017-03-01 Classification: RNA BINDING PROTEIN Ligands: PGE, CA |
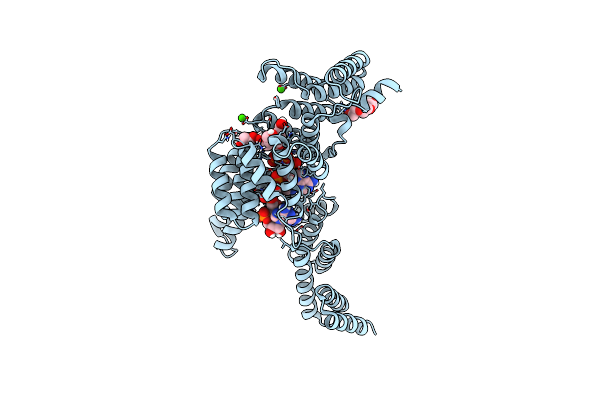 |
Organism: Homo sapiens, Unidentified
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-03-01 Classification: RNA BINDING PROTEIN Ligands: PGE, PEG, EDO, CA |
 |
Ifit1 N216A Monomeric Mutant (L457E/L464E) With M7Gppp-Aaaa (Anti Conformation Of Cap)
Organism: Homo sapiens, Unidentified
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-03-01 Classification: RNA BINDING PROTEIN Ligands: PGE, CA |
 |
Structure Of Human P38Amapk-Arylpyridazinylpyridine Fragment Complex Used In Inhibitor Discovery
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-06-22 Classification: TRANSFERASE Ligands: VVT, GG5, BME |
 |
Crystal Structure Of Chimeric Beta-Lactamase Ctem-19M At 1.2 Angstrom Resolution
Organism: Escherichia coli, Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2015-11-11 Classification: HYDROLASE Ligands: CL, MG |
 |
Crystal Structure Of Chimeric Beta-Lactamase Ctem-19M At 1.1 Angstrom Resolution
Organism: Escherichia coli, Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2015-11-11 Classification: HYDROLASE Ligands: CL, MG |
 |
Crystal Structures Of Chimeric Beta-Lactamase Ctem-19M Showing Different Conformations
Organism: Escherichia coli, Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-08-12 Classification: HYDROLASE Ligands: CL, MG |
 |
Crystal Structures Of Chimeric Beta-Lactamase Ctem-19M Showing Different Conformations
Organism: Escherichia coli, Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2015-08-12 Classification: HYDROLASE Ligands: CL, MG |
 |
Crystal Structure Of P38 Alpha Map Kinase In Complex With A Novel Isoform Selective Drug Candidate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2015-02-25 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: GG5, 3GF, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-10-15 Classification: HYDROLASE Ligands: CL, SO4, GOL |

