Search Count: 16
 |
Alanine-Glyoxylate Aminotransferase 1 (Agt1) From Arabidopsis Thaliana In Presence Of Serine
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-10-23 Classification: TRANSFERASE Ligands: PLP, 3PY |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2019-10-23 Classification: TRANSFERASE Ligands: PLP, FMT, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2006-08-29 Classification: SIGNALING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-08-09 Classification: ELECTRON TRANSPORT |
 |
Crystal Structure Of The C-Terminal Domain Of The Two-Component System Transmitter Protein Nrii (Ntrb)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2004-06-15 Classification: TRANSFERASE |
 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2003-06-10 Classification: TRANSFERASE Ligands: CA |
 |
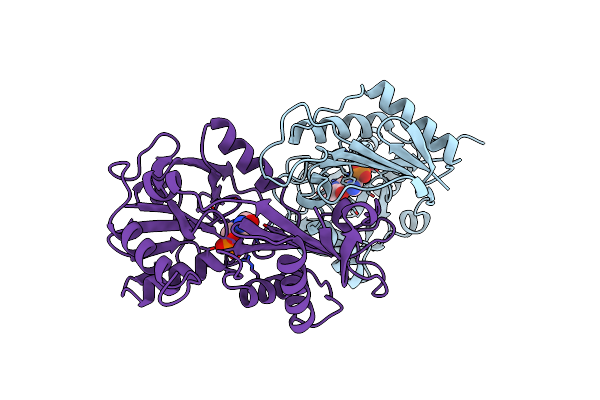
E177K Mutant Of D-Amino Acid Aminotransferase Complexed With Pyridoxamine-5'-Phosphate
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 1998-12-14 Classification: TRANSFERASE Ligands: PLP |
 |
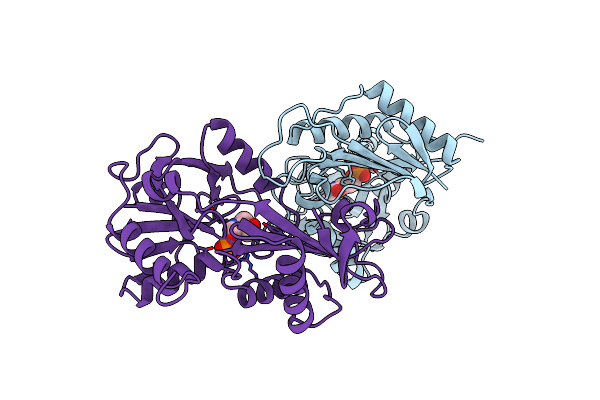
L201A Mutant Of D-Amino Acid Aminotransferase Complexed With Pyridoxamine-5'-Phosphate
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-06-03 Classification: TRANSFERASE Ligands: PMP |
 |
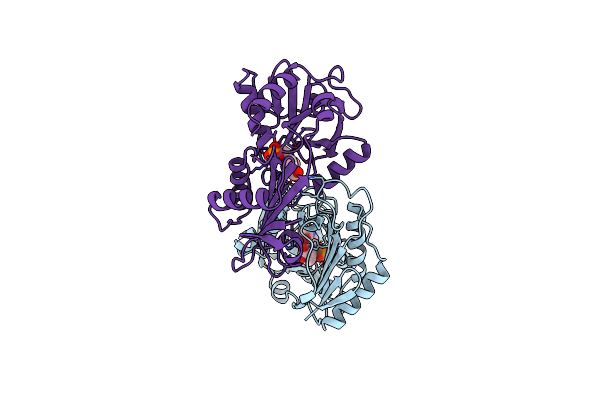
L201A Mutant Of D-Amino Acid Aminotransferase Complexed With Pyridoxal-5'-Phosphate
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-06-03 Classification: TRANSFERASE Ligands: PLP |
 |
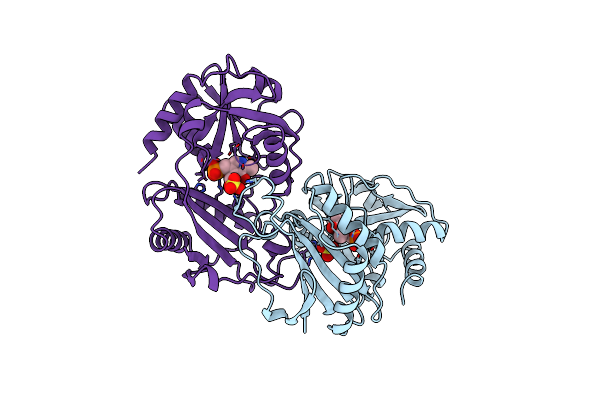
Crystallographic Structure Of D-Amino Acid Aminotransferase Inactivated By Pyridoxyl-D-Alanine
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1998-04-29 Classification: AMINOTRANSFERASE Ligands: PDD |
 |
Crystallographic Structure Of D-Amino Acid Aminotransferase In Pyridoxal-5'-Phosphate (Plp) Form
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1998-04-29 Classification: AMINOTRANSFERASE Ligands: SO4, PLP |
 |
Crystallographic Structure Of D-Amino Acid Aminotransferase Inactivated By D-Cycloserine
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1998-03-18 Classification: PYRIDOXAL PHOSPHATE Ligands: DCS |
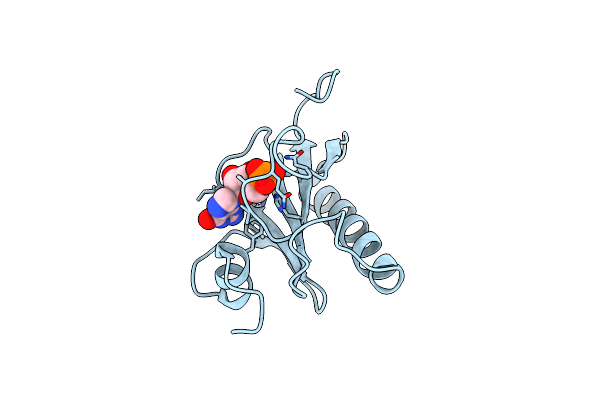 |
Histidine Triad Nucleotide-Binding Protein (Hint) From Rabbit Complexed With Gmp
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1997-06-16 Classification: NUCLEOTIDE-BINDING PROTEIN Ligands: 5GP |
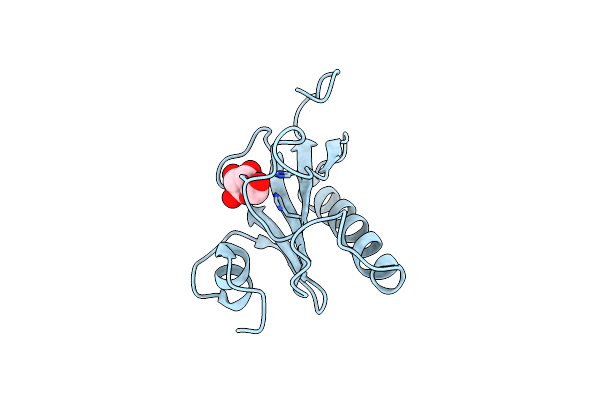 |
Histidine Triad Nucleotide-Binding Protein (Hint) From Rabbit Complexed With Adenosine
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1997-06-16 Classification: NUCLEOTIDE-BINDING PROTEIN Ligands: RIB |
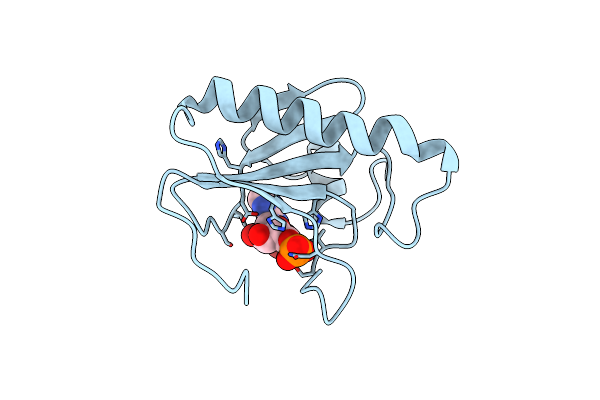 |
Histidine Triad Nucleotide-Binding Protein (Hint) From Rabbit Complexed With 8-Br-Amp
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 1997-06-16 Classification: NUCLEOTIDE-BINDING PROTEIN Ligands: 8BR |
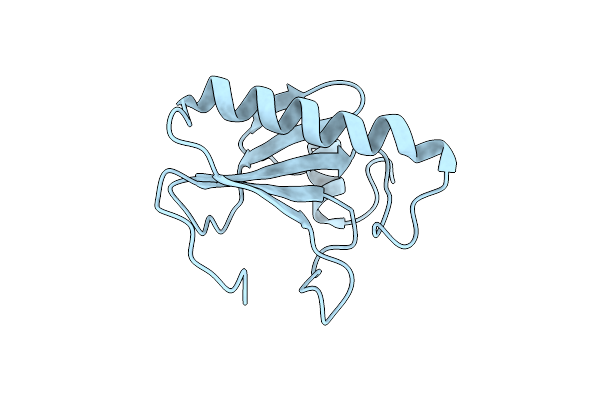 |
Histidine Triad Nucleotide-Binding Protein (Hint) From Rabbit Without Nucleotide
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 1997-06-16 Classification: NUCLEOTIDE-BINDING PROTEIN |

