Search Count: 15
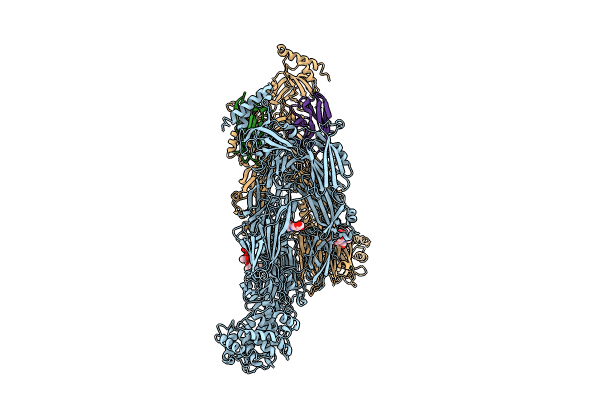 |
Organism: Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-05-04 Classification: IMMUNE SYSTEM Ligands: CA, NAG |
 |
Organism: Lama glama
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2022-03-23 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Organism: Lama glama, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-02-16 Classification: IMMUNE SYSTEM Ligands: NAG |
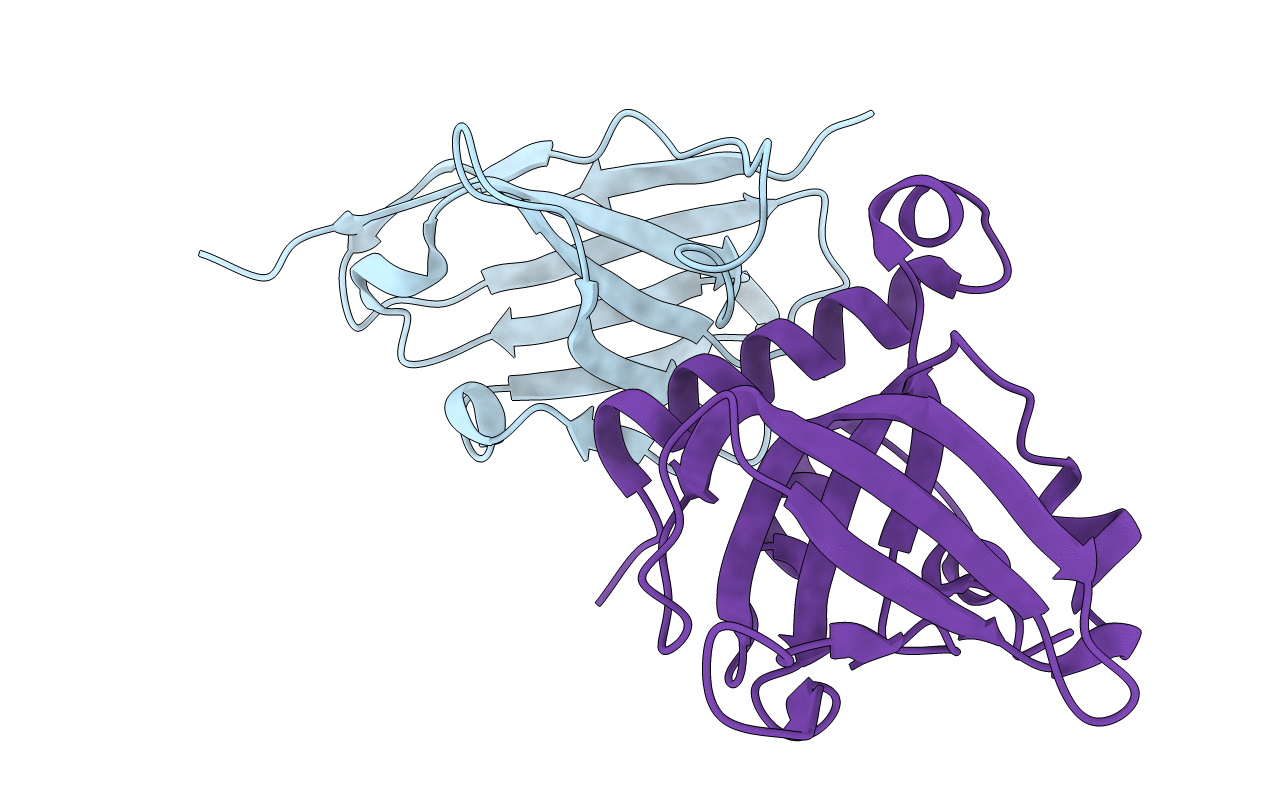 |
Complex Of C-Terminal Domain Of Murine Complement C3B With The Hc3Nb3 Nanobody
Organism: Lama glama, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-08-12 Classification: IMMUNE SYSTEM |
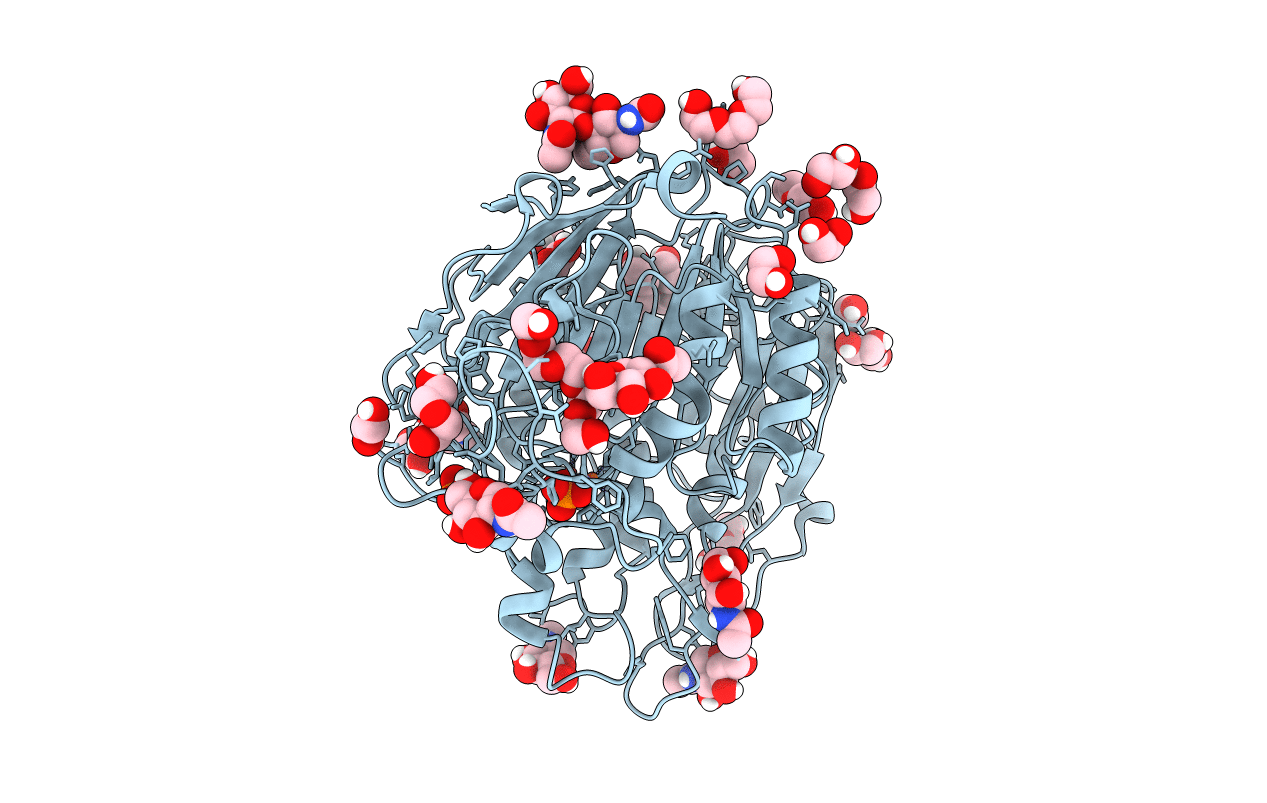 |
Organism: Triticum aestivum
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2019-11-27 Classification: HYDROLASE Ligands: FE, NAG, PO4, PEG, PGE, 1PE, EDO |
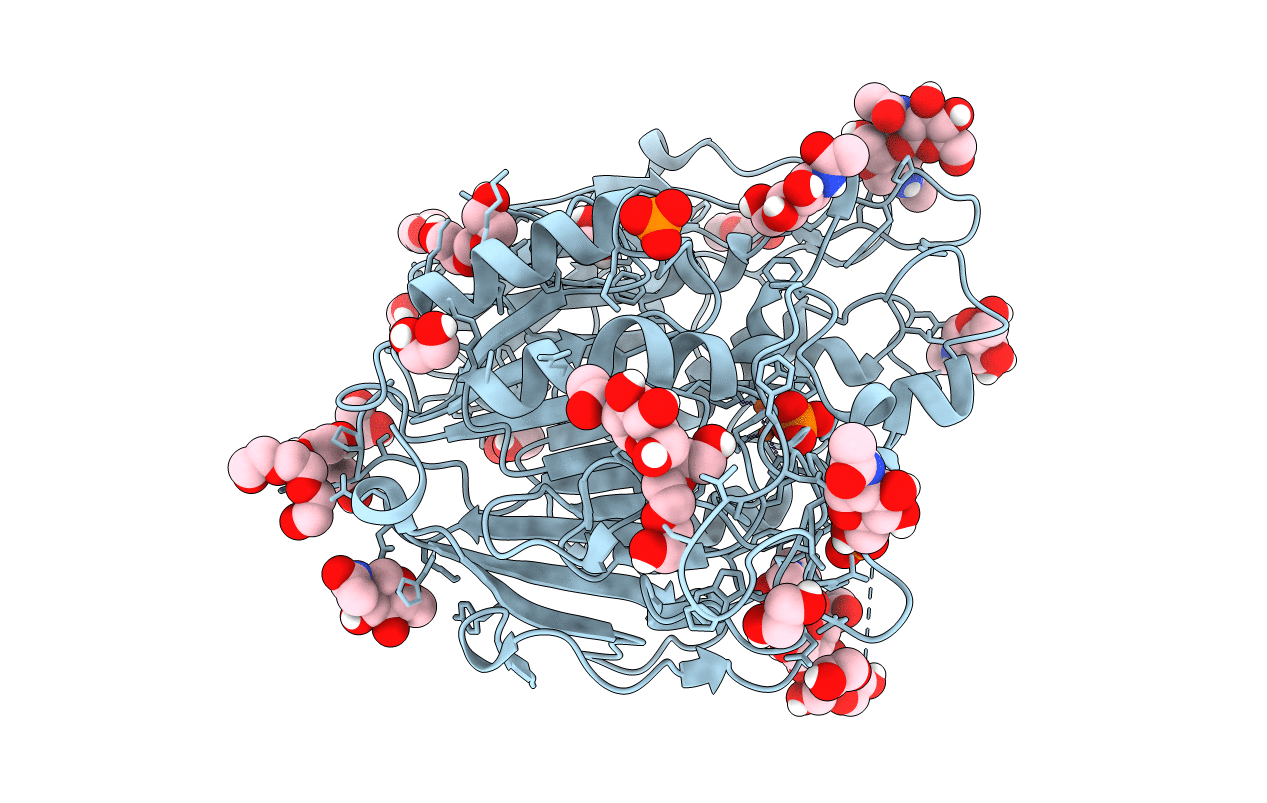 |
Organism: Triticum aestivum
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2019-11-27 Classification: HYDROLASE Ligands: FE, NAG, PO4, PGE, PEG, EDO |
 |
Purple Acid Phytase From Wheat Isoform B2 - Complex With Inositol Hexasulphate
Organism: Triticum aestivum
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2019-11-27 Classification: HYDROLASE Ligands: FE, NAG, PEG, PGE, PO4, PG4, EDO, IHS |
 |
Organism: Triticum aestivum
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2019-11-27 Classification: HYDROLASE Ligands: FE, NAG, PO4, EDO, PEG |
 |
Organism: Triticum aestivum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-11-27 Classification: HYDROLASE Ligands: FE, NAG, PO4, PEG, EDO, PGE, PG6 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2018-12-19 Classification: SUGAR BINDING PROTEIN |
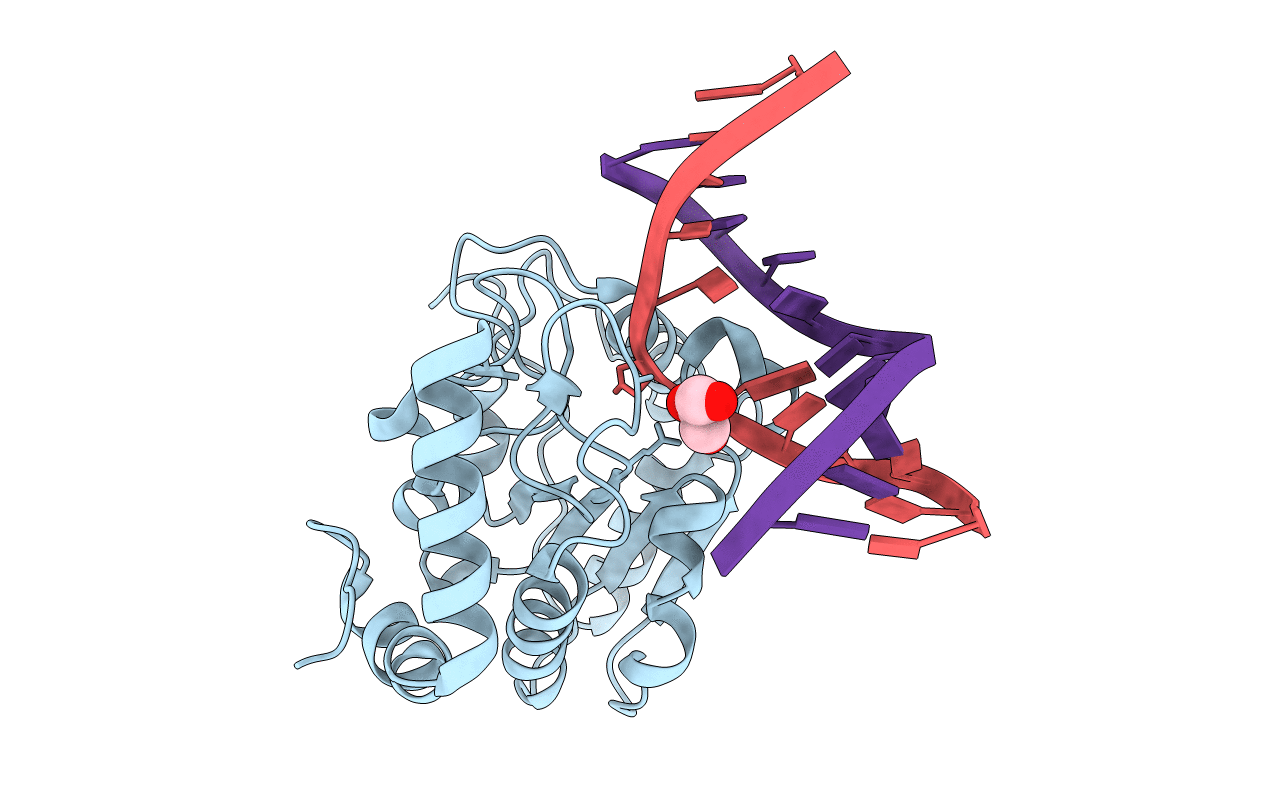 |
Crystal Structure Determination Of Uracil-Dna N-Glycosylase (Ung) From Deinococcus Radiodurans In Complex With Dna - New Insights Into The Role Of The Leucine-Loop For Damage Recognition And Repair
Organism: Deinococcus radiodurans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2015-08-12 Classification: HYDROLASE/DNA Ligands: CL, GOL |
 |
Atomic Resolution Structure Of Tmcbm61 In Complex With Beta-1,4- Galactotriose
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2010-09-08 Classification: HYDROLASE Ligands: CA |
 |
Structure Of Tmcbm61 In Complex With Beta-1,4-Galactotriose At 1.4 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-09-08 Classification: HYDROLASE Ligands: CA, SO4, EDO |
 |
The Crystal Structure Of Iron Superoxide Dismutase From Aliivibrio Salmonicida.
Organism: Aliivibrio salmonicida
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-02-17 Classification: OXIDOREDUCTASE Ligands: FE |

