Search Count: 38
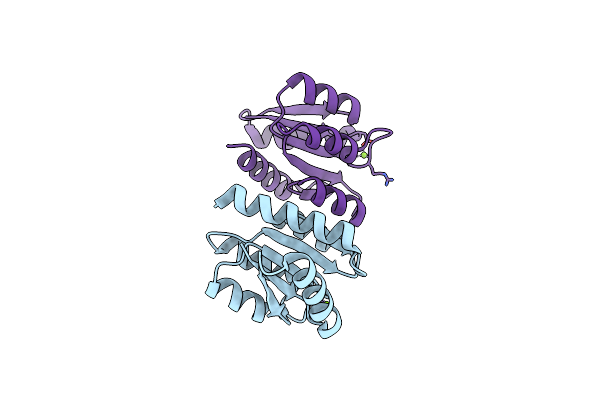 |
The Rec Domain (In The Non-Phosphorylated State) Of Xync, A Response Regulator From G.Proteiniphilus T-6
Organism: Geobacillus proteiniphilus
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: DNA BINDING PROTEIN Ligands: MG |
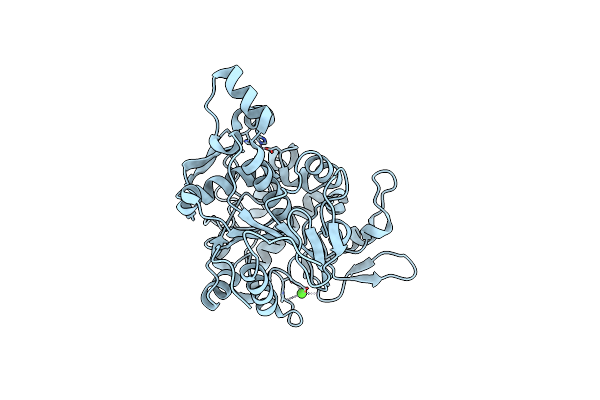 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Variant A187C/F291C
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-10-09 Classification: HYDROLASE Ligands: ZN, CA |
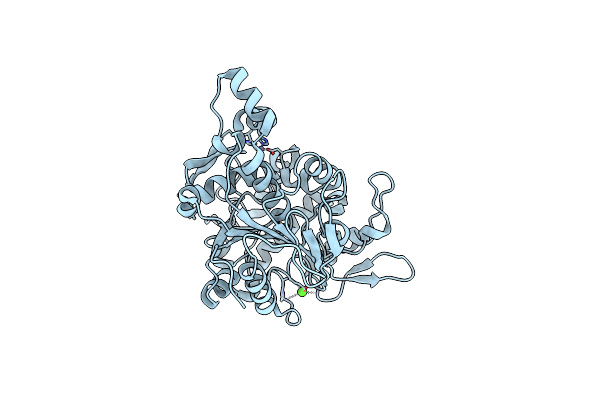 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Variant E134C/F149C
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-10-09 Classification: HYDROLASE Ligands: ZN, CA |
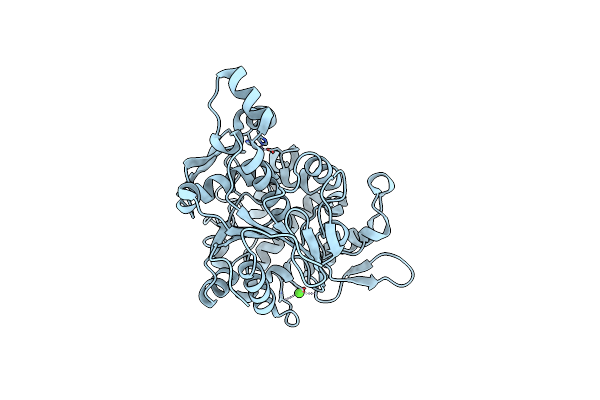 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Methanol Stable Variant E251C/G332C
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-10-09 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Tyrosinase From Bacillus Megaterium With Jkb Inhibitor In The Active Site.
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-06-19 Classification: OXIDOREDUCTASE Ligands: CU, JKB |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Methanol Stable Variant L360F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: CA, ZN |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Methanol Stable Variant L184F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Methanol Stable Variant L184F/A187F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Methanol Stable Variant A187F/L360F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: CA, ZN |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Methanol Stable Variant A187F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Variant L184F/L360F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of Lipase From Geobacillus Stearothermophilus T6 Variant L184F/A187F/L360F
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: ZN, CA |
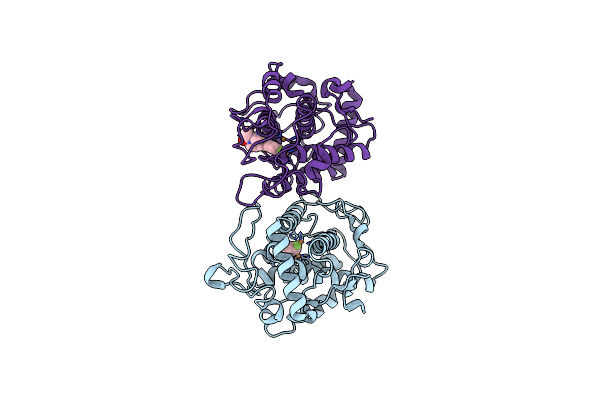 |
Crystal Structure Of Tyrosinase From Bacillus Megaterium With Svf Inhibitor In The Active Site
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-04-25 Classification: OXIDOREDUCTASE Ligands: CU, SVF |
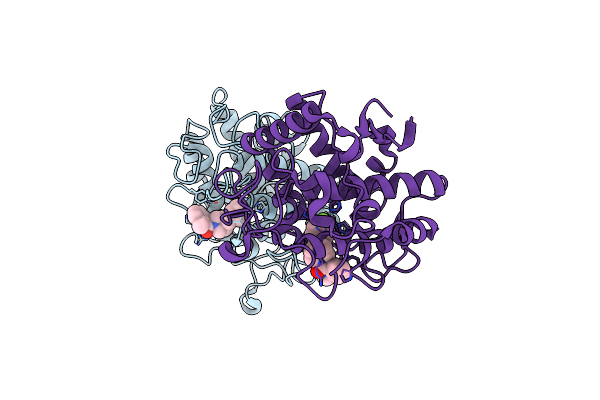 |
Crystal Structure Of Tyrosinase From Bacillus Megaterium With B5N Inhibitor In The Active Site
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-04-25 Classification: OXIDOREDUCTASE Ligands: B5N, CU |
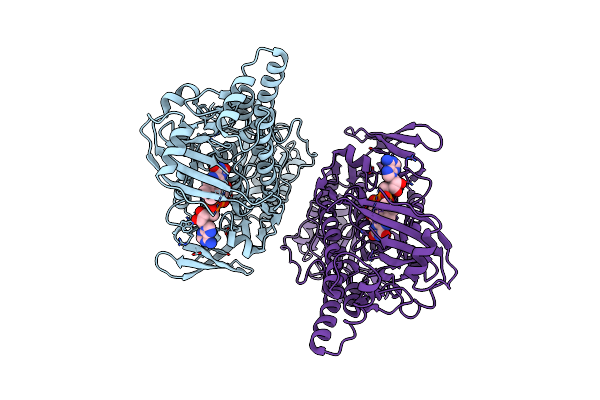 |
Crystal Structure Of 2-Hydroxybiphenyl 3-Monooxygenase M321A From Pseudomonas Azelaica
Organism: Pseudomonas nitroreducens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2018-01-10 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2011-08-31 Classification: SIGNALING PROTEIN Ligands: GOL, MN, NH4, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-08-31 Classification: SIGNALING PROTEIN Ligands: MG |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-05-11 Classification: SIGNALING PROTEIN Ligands: BEF, MN, GOL, SO4 |
 |
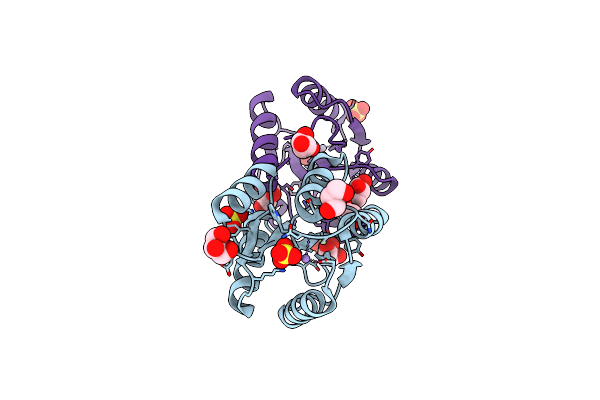
Crystal Structure Of The Chex-Chey-Bef3-Mg+2 Complex From Borrelia Burgdorferi
Organism: Borrelia burgdorferi
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2010-02-16 Classification: SIGNALING PROTEIN Ligands: MG |
 |
Crystal Structure Of Chey Triple Mutant F14E, N59M, E89L Complexed With Bef3- And Mn2+
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-09-22 Classification: SIGNALING PROTEIN Ligands: MN, BEF, GOL, SO4 |

