Search Count: 16
 |
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2025-06-18 Classification: RNA BINDING PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2025-06-18 Classification: RNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: HYDROLASE Ligands: A1IIH, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: A1H35, EDO |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: MEMBRANE PROTEIN |
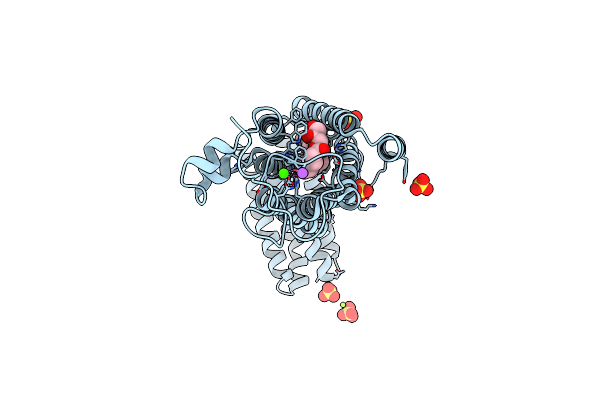 |
Crystal Structure Of Human Alkaline Ceramidase 3 (Acer3) At 2.7 Angstrom Resolution
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-01-02 Classification: MEMBRANE PROTEIN Ligands: ZN, SO4, NA, OLB, CA, MG |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2016-05-04 Classification: DE NOVO PROTEIN Ligands: 60C |
 |
Crystal Structure De Novo Designed Fullerene Organizing Protein Complex With Fullerene
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-05-04 Classification: DE NOVO PROTEIN Ligands: 60C |
 |
Crystal Structure Of De Novo Designed Fullerene Organising Protein Complex With Fullerene
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2016-05-04 Classification: DE NOVO PROTEIN Ligands: 60C |
 |
Organism: Pseudomonas sp. lz4w
Method: SOLUTION NMR Release Date: 2016-01-27 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of The Complex Of Bovine Lactoperoxidase With Carbon Monoxide At 2.0 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-02-15 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, BR, MPD, EDO, SCN, CMO |
 |
The Crystal Structure Of Glycogen Phosphorylase In Complex With Glucose At 100 K
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2008-04-01 Classification: TRANSFERASE Ligands: GLC, DMS |
 |
Crystal Structure Of Glycogen Phosphorylase In Complex With Glucosyl Triazoleacetamide
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2008-04-01 Classification: TRANSFERASE Ligands: DL8, DMS |

