Search Count: 20
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:3.42 Å Release Date: 2020-06-10 Classification: GENE REGULATION Ligands: IPH, ZN |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2018-08-01 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: C8Y, CIT, EDO |
 |
Crystal Structure Kpc-2 Beta-Lactamase Complexed With Wck 4234 By Co-Crystallization
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-08-01 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: C8Y, SO4, CL, ACT |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-08-01 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: C8V, CIT, EDO |
 |
Crystal Structure Kpc-2 Beta-Lactamase Complexed With Wck 5107 By Co-Crystallization
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2018-08-01 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: SO4, CL, C8V |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-08-01 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: C9D, CIT, EDO |
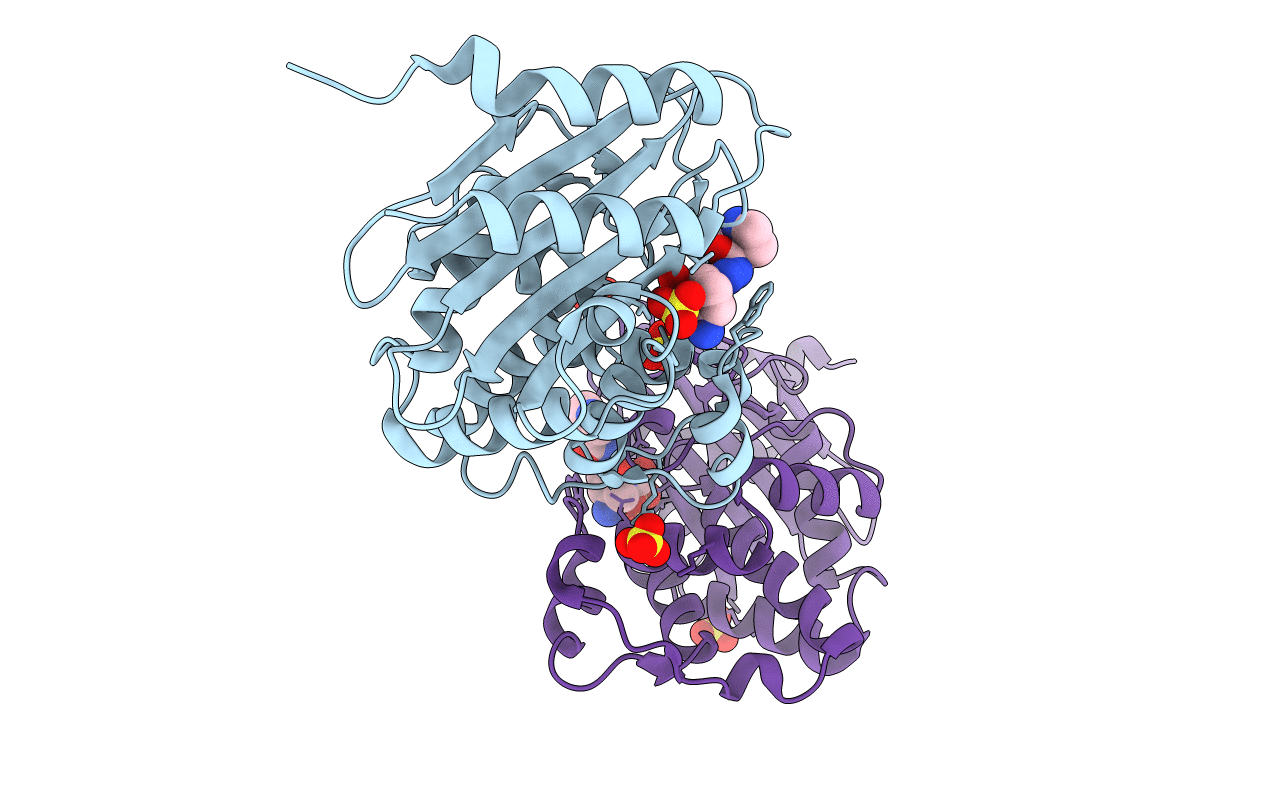 |
Crystal Structure Kpc-2 Beta-Lactamase Complexed With Wck 5153 By Co-Crystallization
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-08-01 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: CD7, SO4, CL |
 |
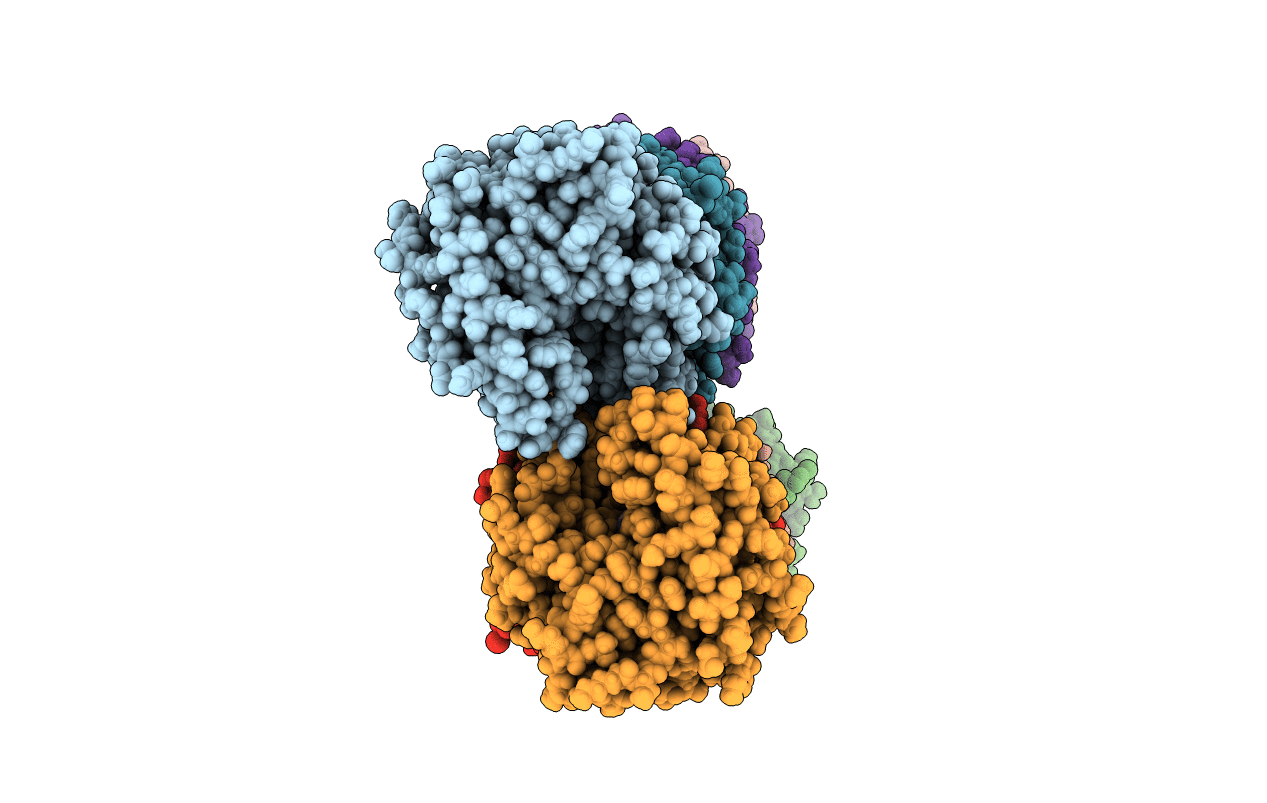
Crystal Structure Oxa-24 Beta-Lactamase Complexed With Wck 4234 By Co-Crystallization
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2018-08-01 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: C8Y, CL |
 |
Crystal Structure Of Uracil Dna Glycosylase-Uracil Complex From Bradyrhizobium Diazoefficiens.
Organism: Bradyrhizobium diazoefficiens usda 110
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2017-06-14 Classification: HYDROLASE Ligands: URA |
 |
Crystal Structure Of Uracil Dna Glycosylase (Bdiung) From Bradyrhizobium Diazoefficiens
Organism: Bradyrhizobium diazoefficiens usda 110
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-05-03 Classification: HYDROLASE |
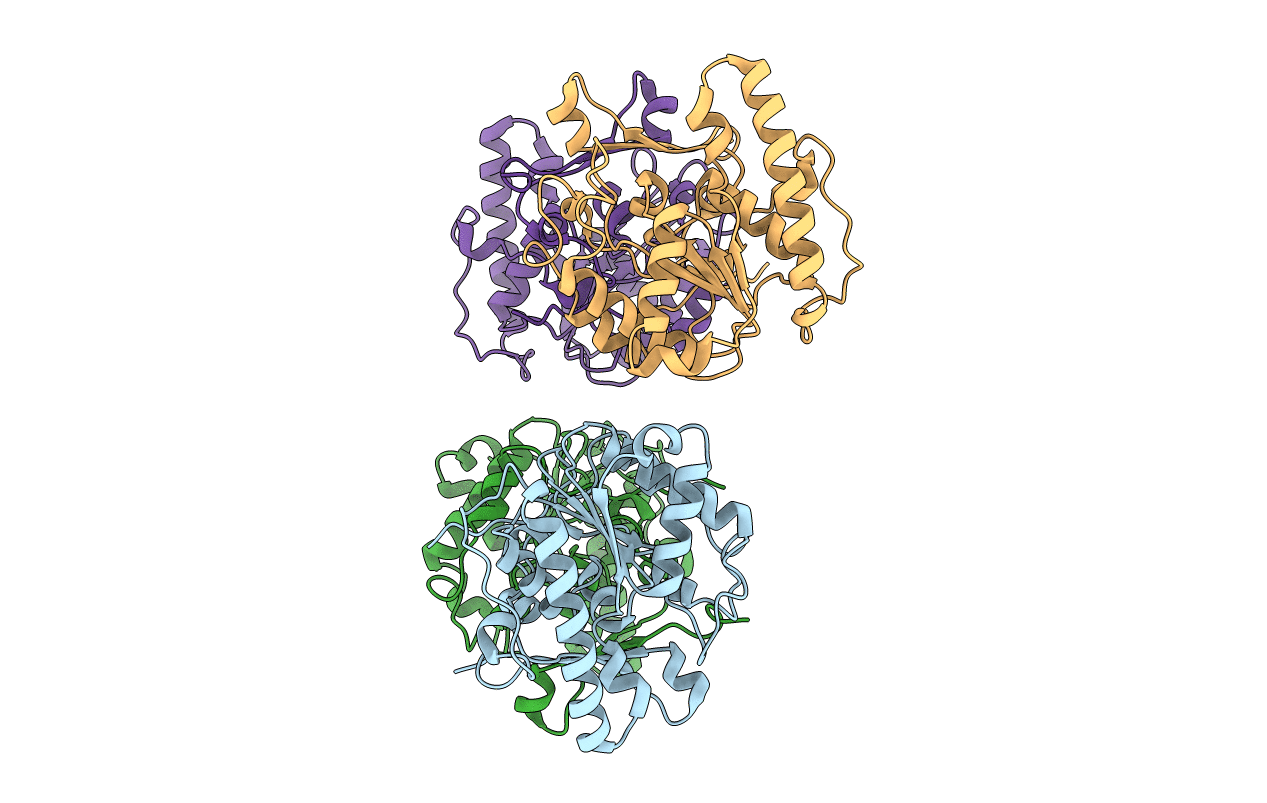 |
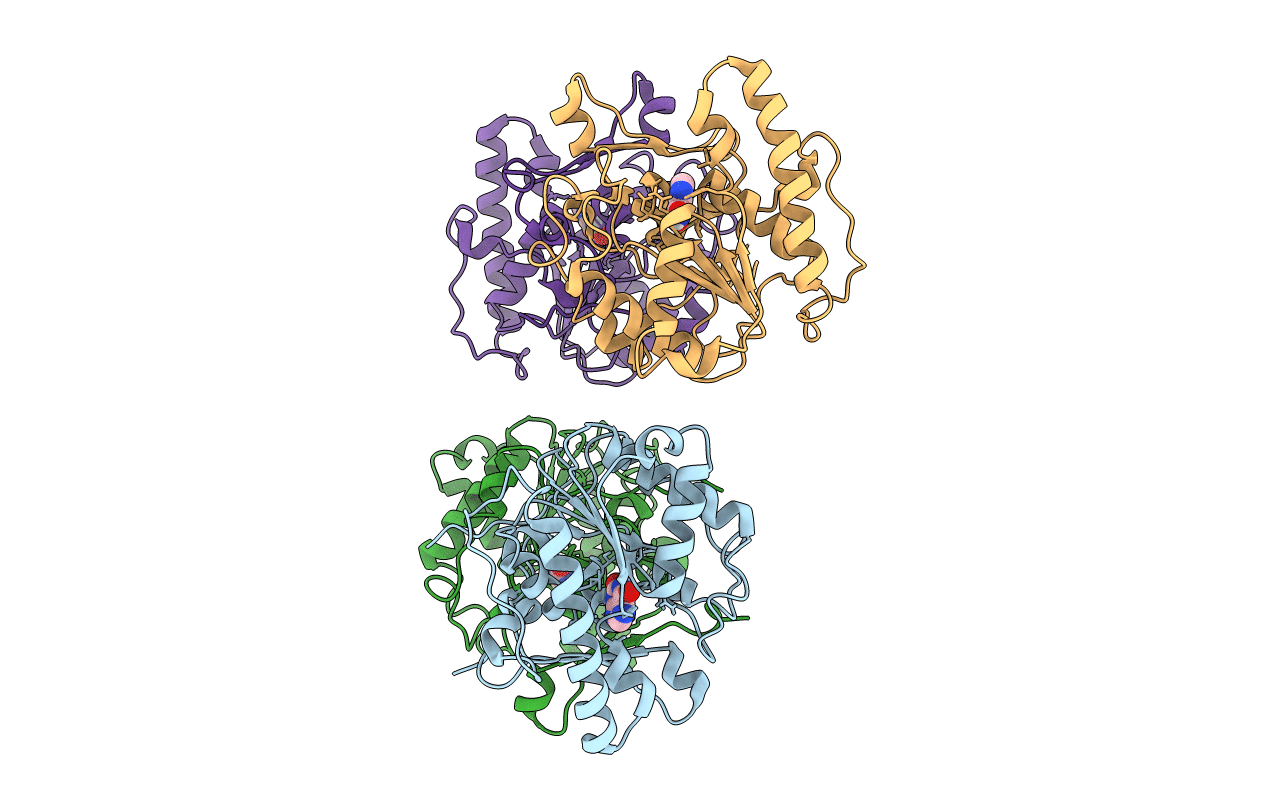
Structure Of Selenomethionine-Labelled Uracil Dna Glycosylase (Bdiung) From Bradyrhizobium Diazoefficiens
Organism: Bradyrhizobium diazoefficiens usda 110
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2017-05-03 Classification: HYDROLASE |
 |
Crystal Structure Of Uracil Dna Glycosylase -Xanthine Complex From Bradyrhizobium Diazoefficiens
Organism: Bradyrhizobium diazoefficiens usda 110
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-05-03 Classification: HYDROLASE Ligands: XAN |
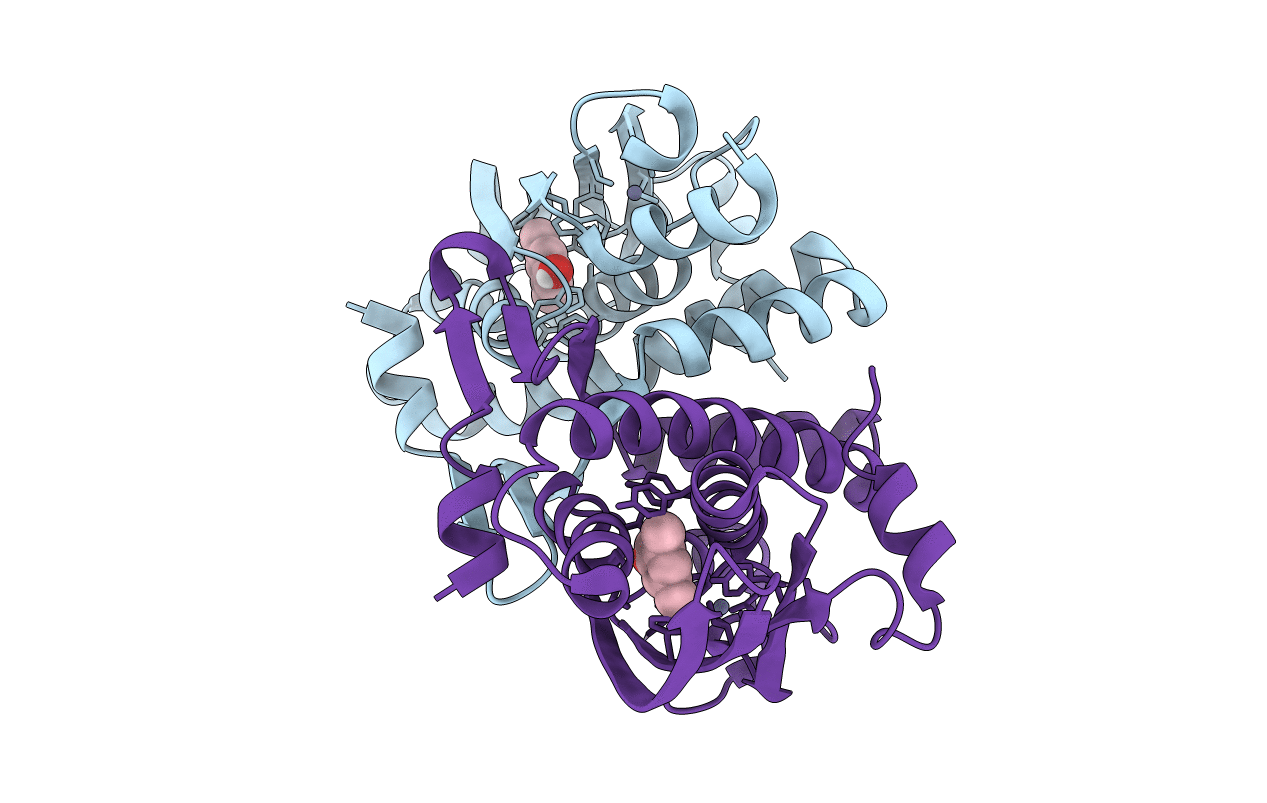 |
Crystal Structure Of The Phenol-Responsive Sensory Domain Of The Transcription Activator Poxr With 3,5-Dimethylphenol
Organism: Ralstonia sp. e2
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2016-05-18 Classification: TRANSCRIPTION Ligands: ZN, ERH |
 |
Crystal Structure Of The Phenol-Responsive Sensory Domain Of The Transcription Activator Poxr With Phenol
Organism: Ralstonia sp. e2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-04-06 Classification: TRANSCRIPTION Ligands: IPH, ZN |
 |
Crystal Structure Of The Phenol-Responsive Sensory Domain Of The Transcription Activator Poxr
Organism: Ralstonia sp. e2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-03-30 Classification: TRANSCRIPTION Ligands: ZN |
 |
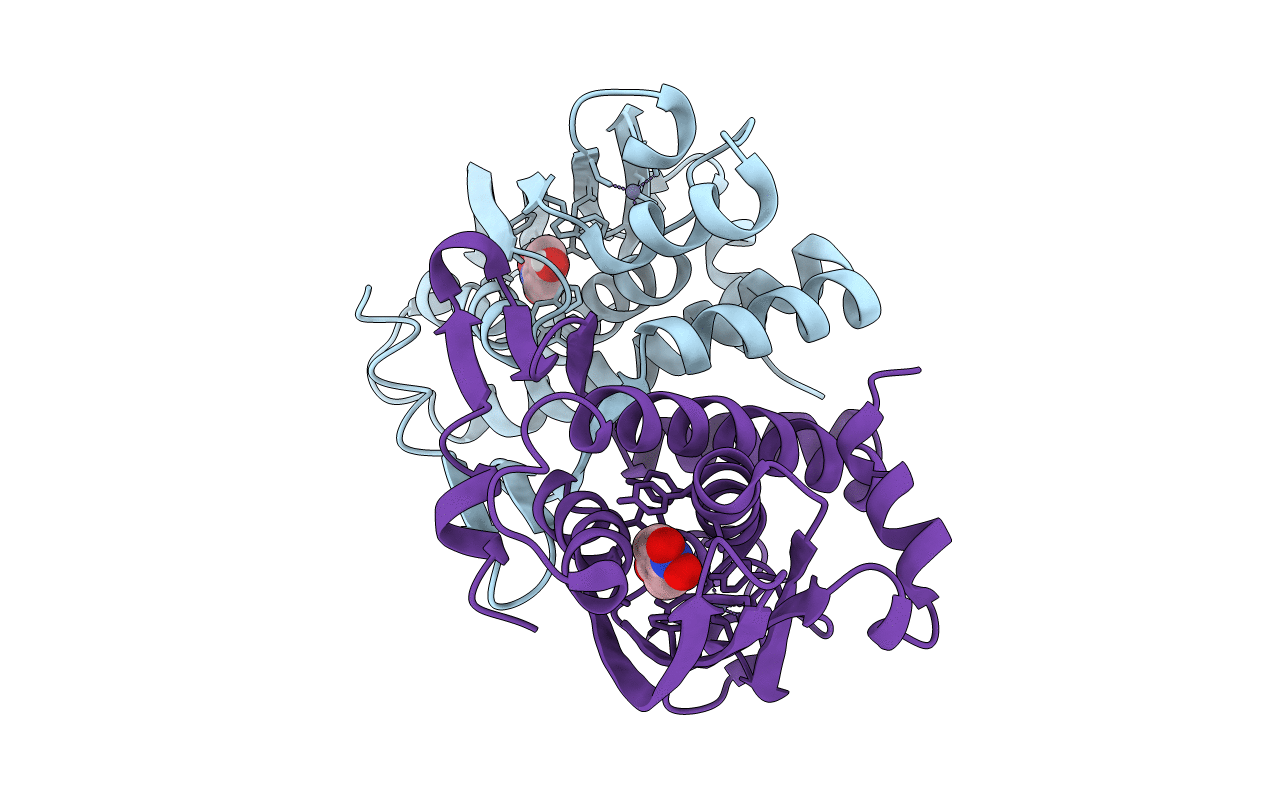
Crystal Structure Of The Phenol-Responsive Sensory Domain Of The Transcription Activator Poxr In Complex With 4-Methylphenol (Cresol)
Organism: Ralstonia sp. e2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-03-30 Classification: TRANSCRIPTION Ligands: ZN, PCR |
 |
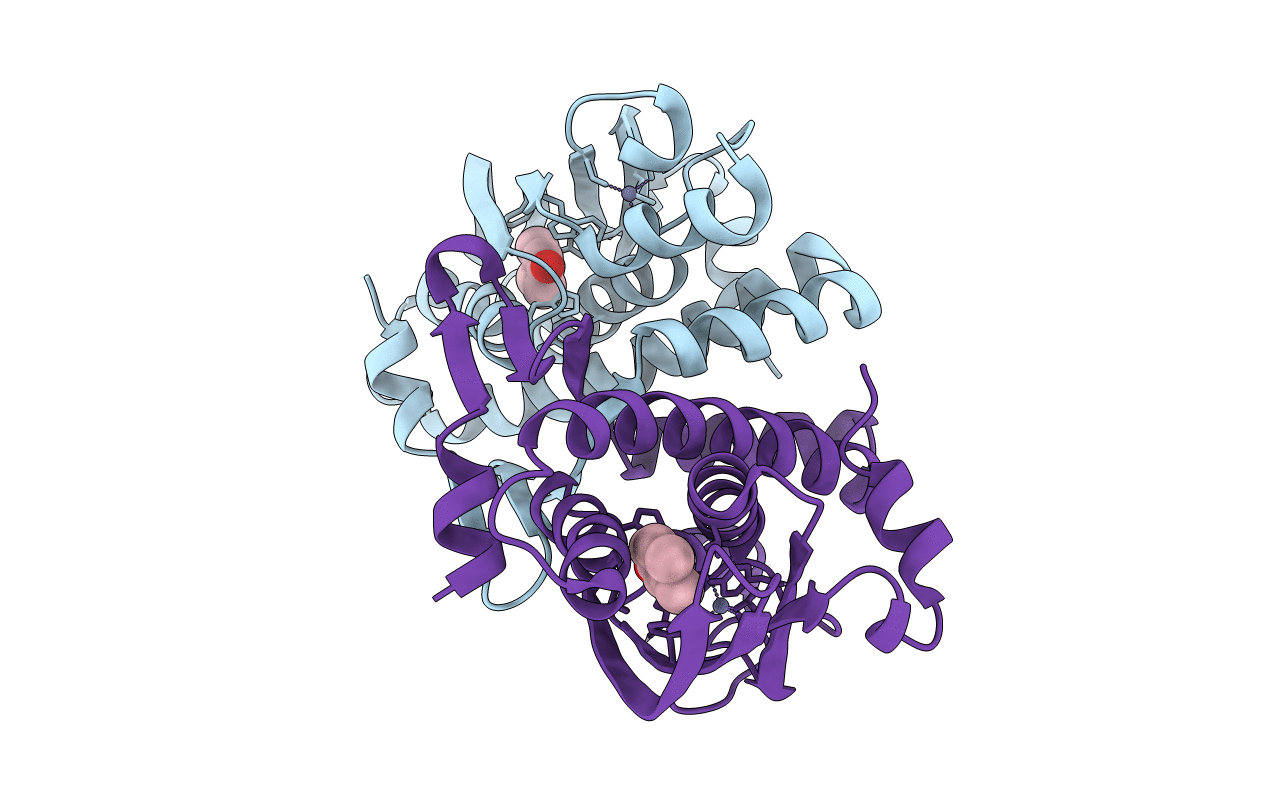
Crystal Structure Of The Phenol-Responsive Sensory Domain Of The Transcription Activator Poxr In Complex With 4-Nitrophenol
Organism: Ralstonia sp. e2
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-03-30 Classification: TRANSCRIPTION Ligands: ZN, NPO |
 |
Crystal Structure Of The Phenol-Responsive Sensory Domain Of The Transcription Activator Poxr With 3,4-Dimethylphenol
Organism: Ralstonia sp. e2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-03-30 Classification: TRANSCRIPTION Ligands: ZN, 2MP |
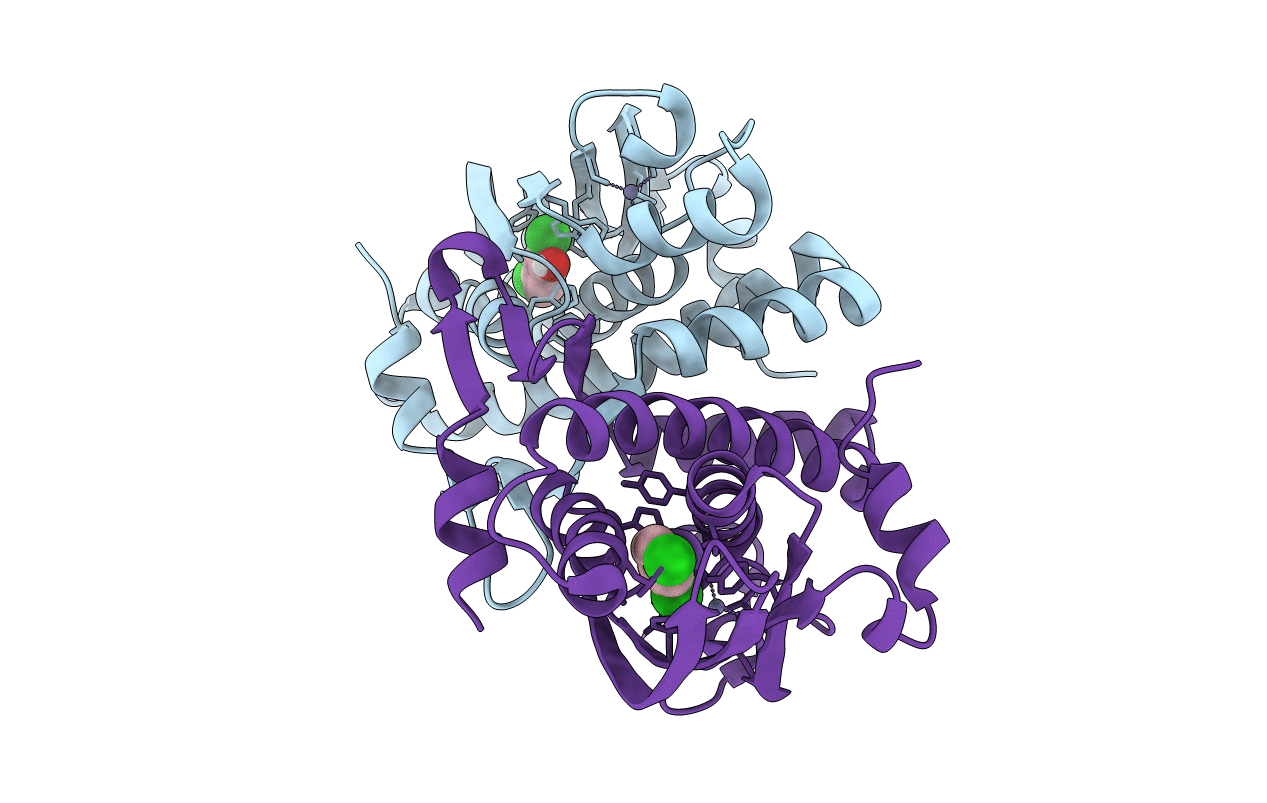 |
Crystal Structure Of The Phenol-Responsive Sensory Domain Of The Transcription Activator Poxr With 2,4-Dichlorophenol
Organism: Ralstonia sp. e2
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-03-30 Classification: TRANSCRIPTION Ligands: 5JC, ZN |
 |
Structure Of The M. Smegmatis Gyrb Atpase Domain In Complex With An Aminopyrazinamide
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-01-23 Classification: ISOMERASE Ligands: B5U, NA |

