Search Count: 19
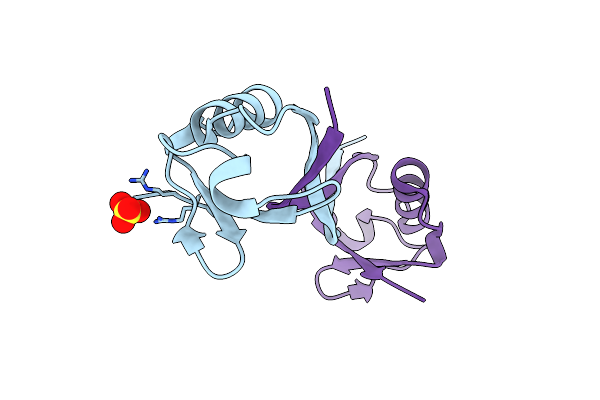 |
Structure Of A Dimeric Ubiquitin Variant (Ubv3) That Inhibits The Protease (Pro) From Turnip Yellow Mosaic Virus (Tymv)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2025-01-22 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
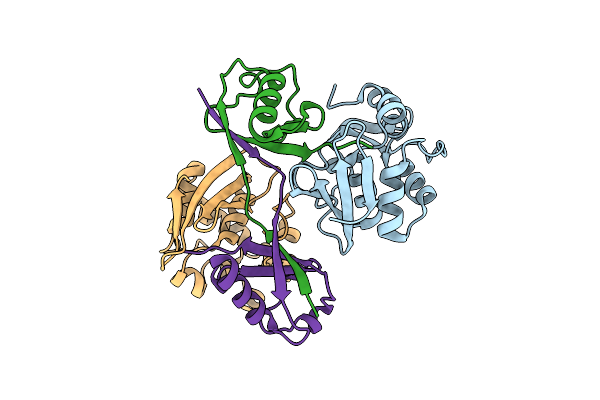 |
Turnip Yellow Mosaic Virus (Tymv) Protease (Pro) Bound To A Ubiquitin Variant (Ubv3)
Organism: Turnip yellow mosaic virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN |
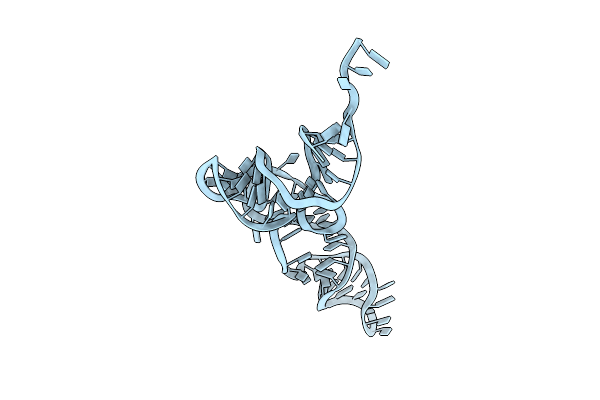 |
Organism: Methanosarcina mazei
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: RNA |
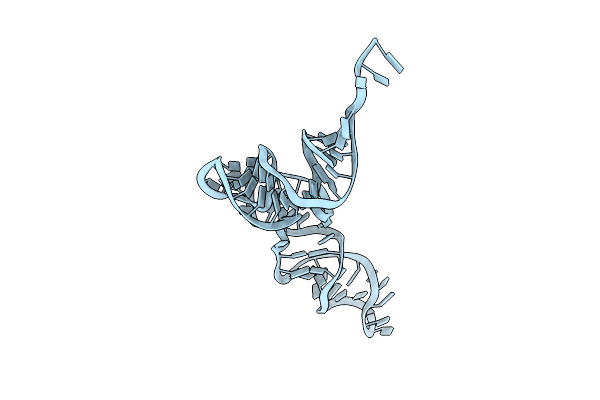 |
Organism: Candidatus methanomethylophilus alvus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: RNA |
 |
Organism: Streptococcus pyogenes serotype m3 (strain atcc baa-595 / mgas315), Streptococcus mutans serotype c (strain atcc 700610 / ua159)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-07-28 Classification: VIRAL PROTEIN/TRANSCRIPTION |
 |
Organism: Streptococcus pyogenes serotype m3 (strain atcc baa-595 / mgas315), Streptococcus mutans serotype c (strain atcc 700610 / ua159)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-07-28 Classification: VIRAL PROTEIN/TRANSCRIPTION Ligands: EPE |
 |
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni, N84A Mutant With Pyruvate Bound In The Active Site
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-12-04 Classification: LYASE Ligands: MG, EDO, ACT, PGE, PEG, GOL |
 |
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni, N84D Mutant With Pyruvate Bound In The Active Site
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2019-12-04 Classification: LYASE Ligands: EDO, PG4, MG, PGE, ACT, GOL, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2017-11-22 Classification: DNA Ligands: K |
 |
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2017-05-24 Classification: ISOMERASE/TRANSFERASE Ligands: CA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2016-02-10 Classification: Laminin Binding Protein Ligands: NAG, CA, TAM |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2015-02-25 Classification: APOPTOSIS Ligands: NAG, CA, CL |
 |
Ampr Effector Binding Domain From Citrobacter Freundii Bound To Udp-Murnac-Pentapeptide
Organism: Citrobacter freundii atcc 8090 = mtcc 1658
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-12-17 Classification: TRANSCRIPTION Ligands: GOL, MUB |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2014-01-01 Classification: TRANSFERASE Ligands: ACP |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-01-01 Classification: TRANSFERASE Ligands: AH0, ACP, MG |
 |
Organism: Synechococcus elongatus pcc 6301
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2009-11-24 Classification: ISOMERASE Ligands: PXG |
 |
Organism: Synechococcus elongatus pcc 6301
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-11-24 Classification: ISOMERASE Ligands: PMP |
 |
Organism: Synechococcus elongatus pcc 6301
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2009-11-24 Classification: ISOMERASE Ligands: PMP, GAB |
 |
Method: X-RAY DIFFRACTION
Resolution:1.83 Å Release Date: 2009-05-05 Classification: STRUCTURAL PROTEIN |

