Search Count: 22
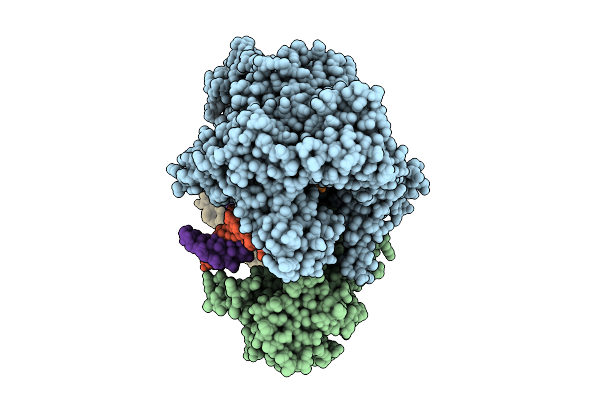 |
Cryo-Em Structure Of The Human Mitochondrial Rna Polymerase Transcription Initiation Complex (Polrmt/Tfam/Tfb2M/Dna/Rna) With A 2-Mer Rna (Pppgpa) And Gtp Poised For Catalysis (Pre-Ic3)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSCRIPTION Ligands: GTP, MG |
 |
Cryo-Em Structure Of The Human Mitochondrial Rna Polymerase Transcription Initiation Complex (Polrmt/Tfb2M/Dna/Rna) Without Tfam; And With A 2-Mer Rna (Pppgpa) And Gtp Poised For Catalysis (Pre-Ic3-Tfam)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSCRIPTION Ligands: GTP, MG |
 |
Cryo-Em Structure Of The Human Mitochondrial Rna Polymerase Transcription Initiation Complex (Polrmt/Tfb2M/Dna/Rna) Without Tfam; And With A Slipped 3-Mer Rna, Pppgpgpa (Slipped Ic3-Tfam)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSCRIPTION |
 |
Cryo-Em Structure Of The Human Mitochondrial Rna Polymerase Transcription Initiation Complex (Polrmt/Tfam/Tfb2M/Dna/Rna) With A Slipped 3-Mer Rna, Pppgpgpa (Slipped Ic3)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSCRIPTION |
 |
Cryo-Em Structure Of The Human Mitochondrial Rna Polymerase Transcription Initiation Complex (Polrmt/Tfam/Tfb2M/Dna/Rna) With A Slipped 3-Mer Rna (Pppgpgpa) And Gtp Poised For Catalysis (Slipped Pre-Ic4)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSCRIPTION Ligands: GTP, MG |
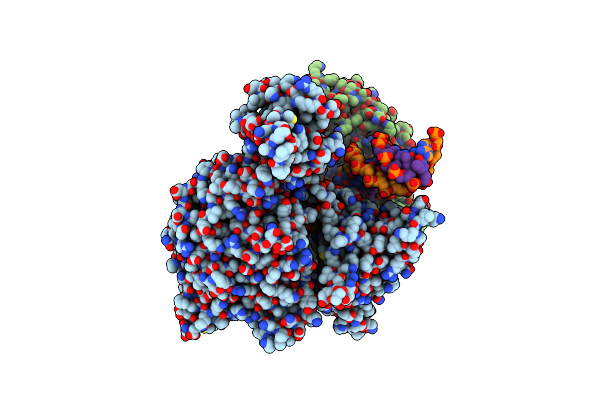 |
Cryo-Em Structure Of Yeast Mitochondrial Rna Polymerase Transcription Initiation Complex With 4-Mer Rna, Pppgpgpupa (Ic4)
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSCRIPTION Ligands: GTP |
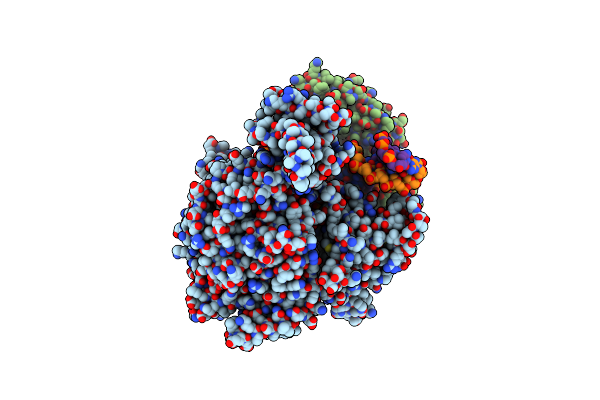 |
Cryo-Em Structure Of Yeast Mitochondrial Rna Polymerase Transcription Initiation Complex With 5-Mer Rna, Pppgpgpapapa (Ic5)
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSCRIPTION Ligands: GTP |
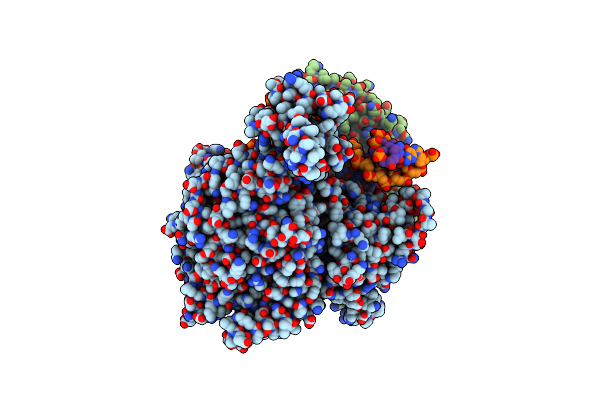 |
Cryo-Em Structure Of Yeast Mitochondrial Rna Polymerase Transcription Initiation Complex With 6-Mer Rna, Pppgpgpapapapu (Ic6)
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSCRIPTION Ligands: GTP |
 |
Cryo-Em Structure Of Yeast Mitochondrial Rna Polymerase Transcription Initiation Complex With 7-Mer Rna, Pppgpgpupapapapu (Ic7)
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSCRIPTION Ligands: GTP |
 |
Cryo-Em Structure Of Yeast Mitochondrial Rna Polymerase Transcription Initiation Complex With 8-Mer Rna, Pppgpgpupapapapupg (Ic8)
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSCRIPTION Ligands: GTP |
 |
Cryo-Em Structure Of Ic8', A Second State Of Yeast Mitochondrial Rna Polymerase Transcription Initiation Complex With 8-Mer Rna, Pppgpgpupapapapupg
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSCRIPTION Ligands: GTP |
 |
Cryo-Em Structure Of Yeast Mitochondrial Rna Polymerase Transcription Initiation Complex With Two Gtp Molecules Poised For De Novo Initiation (Ic2)
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-08-23 Classification: TRANSCRIPTION Ligands: MG, GTP |
 |
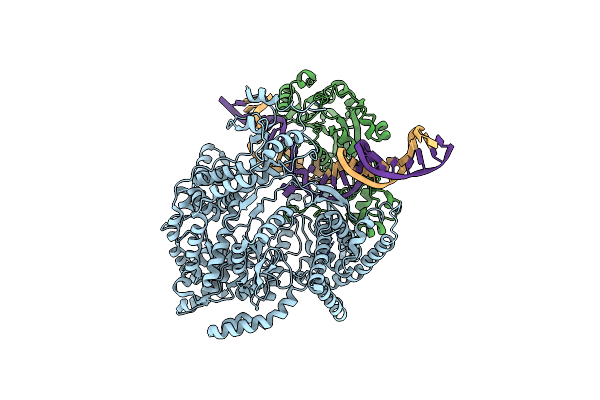
Cryo-Em Structure Of Yeast Mitochondrial Rna Polymerase Transcription Initiation Complex
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae s288c, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: TRANSCRIPTION Ligands: P5E, MG |
 |
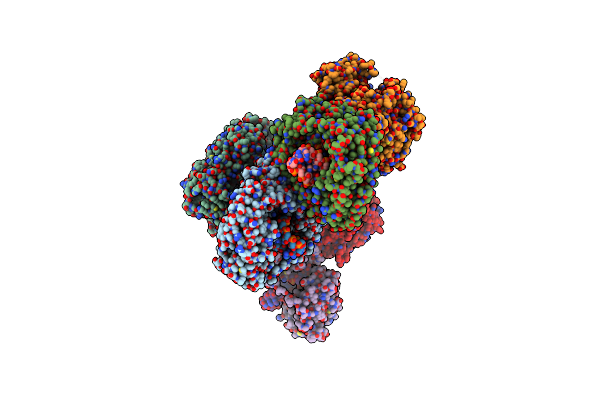
Cryo-Em Structure Of Yeast Mitochondrial Rna Polymerase Partially-Melted Transcription Initiation Complex (Pmic)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-12-02 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.28 Å Release Date: 2016-01-13 Classification: hydrolase/rna Ligands: ZN, MG, M7G |
 |
Crystal Structure Of Rig-I Helicase-Rd In Complex With 24-Mer Blunt-End Hairpin Rna
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-01-13 Classification: hydrolase/rna Ligands: ZN, MG, ETF, BU3 |
 |
Crystal Structure Of Rig-I Helicase-Rd In Complex With 24-Mer 5' Triphosphate Hairpin Rna
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-01-13 Classification: hydrolase/rna Ligands: ZN, MG, GTP |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-11-18 Classification: HYDROLASE/RNA Ligands: ZN, MG, GOL, ADP, BEF |
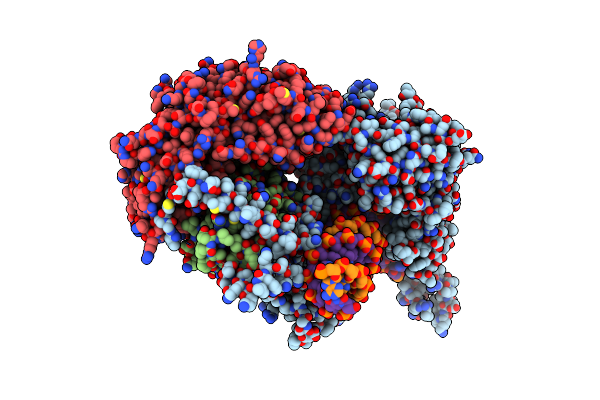 |
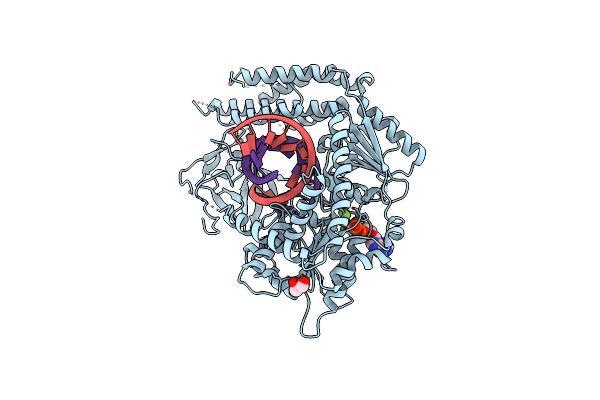
Structural Basis For Processivity And Antiviral Drug Toxicity In Human Mitochondrial Dna Replicase
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2015-09-23 Classification: DNA Binding Protein/DNA Ligands: MG, DCT |
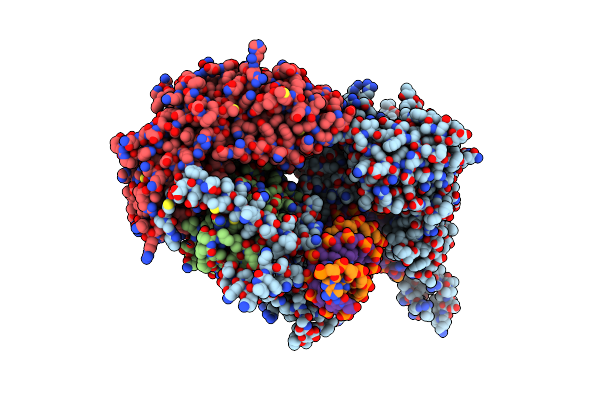 |
Structural Basis For Processivity And Antiviral Drug Toxicity In Human Mitochondrial Dna Replicase
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.44 Å Release Date: 2015-09-23 Classification: DNA Binding Protein/DNA Ligands: MG, DCP |

