Search Count: 17
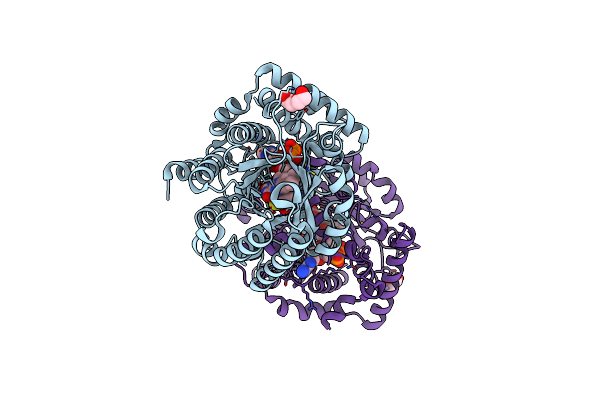 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Complexed With (Prop-2-Ynylthio)Acetic Acid
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, X79 |
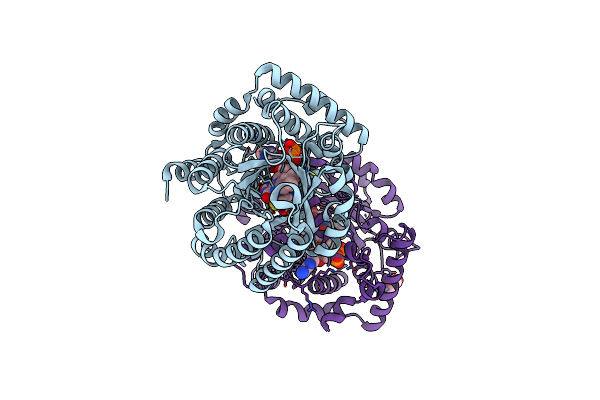 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Complexed With 2-(Cyanomethylthio)Acetic Acid
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FAD, X7K, PEG |
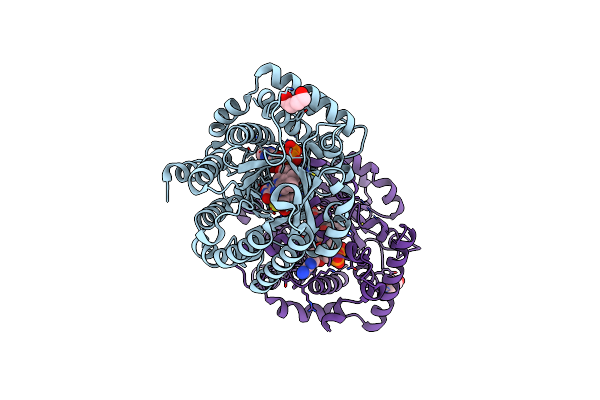 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Complexed With (Allylthio)Acetic Acid
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, X7Q |
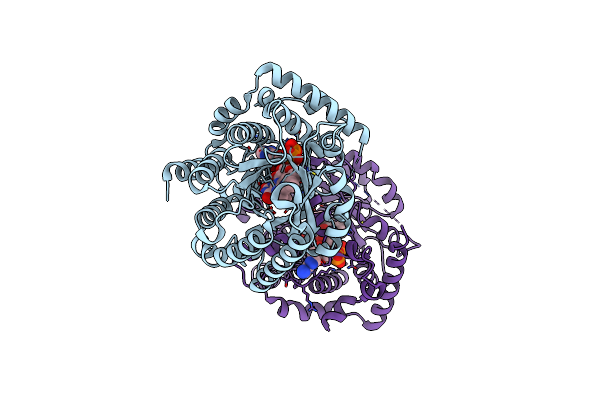 |
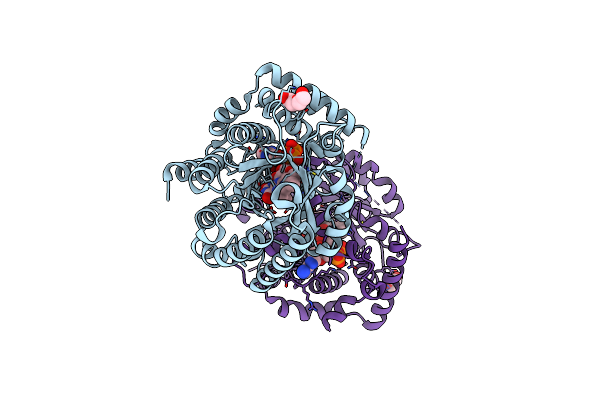
Minimal Puta Proline Dehydrogenase Domain (Design #2) With The Fad N5 Modified With Propanal Resulting From Inactivation With N-Allylglycine(Replicate #1)
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FDA, FAD, CBG |
 |
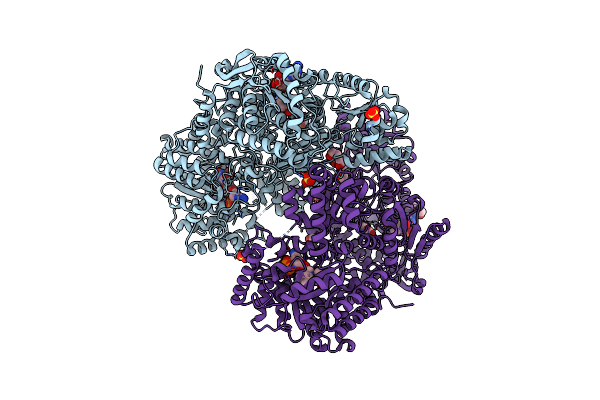
Minimal Puta Proline Dehydrogenase Domain (Design #2) With The Fad N5 Modified With Propanal Resulting From Inactivation With N-Allylglycine (Replicate #2)
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FDA, FAD, CBG, PEG |
 |
Structure Of Proline Utilization A With The Fad Covalently-Modified By Propanal Resulting From Inactivation With N-Allylglycine
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FDA, CBG, NAD, MG, SO4, PEG, A1AV8, PGE |
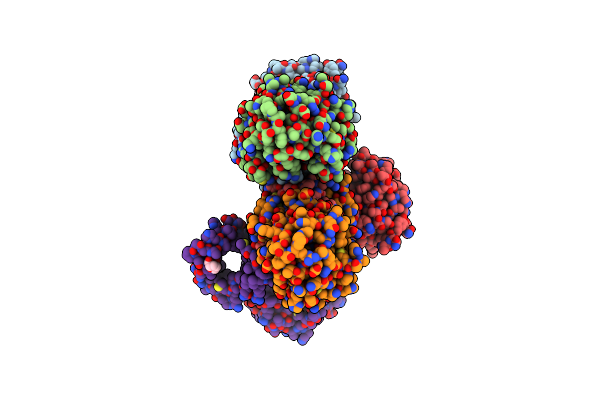 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-11-04 Classification: OXIDOREDUCTASE Ligands: TFB, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-11-04 Classification: OXIDOREDUCTASE Ligands: FPK |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-11-04 Classification: OXIDOREDUCTASE Ligands: T2C, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-11-04 Classification: OXIDOREDUCTASE Ligands: PRS, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-11-04 Classification: OXIDOREDUCTASE Ligands: SO4, IQ0 |
 |
Crystal Structure Of An Sdr From Burkholderia Ambifaria In Complex With Nadph With A Tcep Adduct
Organism: Burkholderia ambifaria (strain mc40-6)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-05-24 Classification: OXIDOREDUCTASE Ligands: 9G1, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2017-03-15 Classification: OXIDOREDUCTASE Ligands: SO4, NDP, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-03-15 Classification: OXIDOREDUCTASE Ligands: PRO, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-03-15 Classification: OXIDOREDUCTASE Ligands: NDP, TFB, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-03-15 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-03-15 Classification: OXIDOREDUCTASE Ligands: CL |

