Search Count: 14
 |
Nmr Shows Why A Small Chemical Change Almost Abolishes The Antimicrobial Activity Of Gccf
Organism: Lactiplantibacillus plantarum
Method: SOLUTION NMR Release Date: 2023-07-05 Classification: ANTIMICROBIAL PROTEIN Ligands: NAG |
 |
Organism: Lactobacillus plantarum
Method: SOLUTION NMR Release Date: 2014-12-10 Classification: ANTIMICROBIAL PROTEIN Ligands: NAG |
 |
Organism: Lactobacillus plantarum
Method: SOLUTION NMR Release Date: 2011-05-04 Classification: ANTIMICROBIAL PROTEIN Ligands: NAG |
 |
Truncated Form Of 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase From Thermotoga Maritima
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-01-26 Classification: TRANSFERASE Ligands: GOL, AZI |
 |
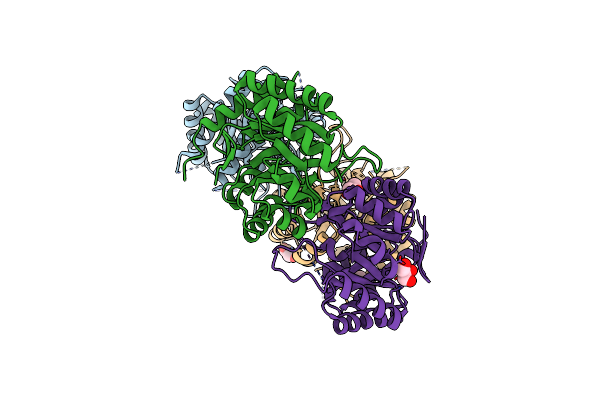
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2011-01-26 Classification: Transferase/Transferase inhibitor Ligands: TYR, AZI, NO3, CL |
 |
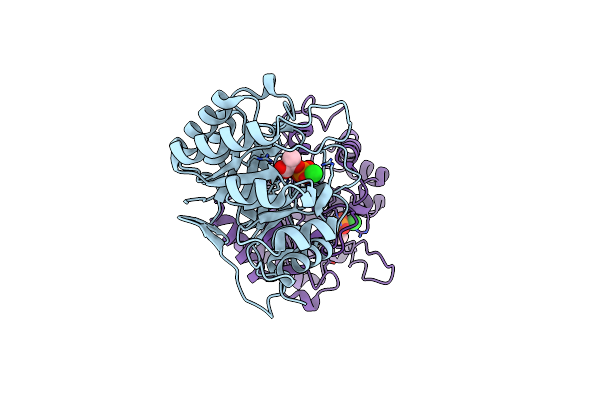
Crystal Structure Of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Synthase (Kdo8Ps) From Neisseria Meningitidis
Organism: Neisseria meningitidis serogroup b
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2008-07-01 Classification: TRANSFERASE Ligands: CL, NA, GOL |
 |
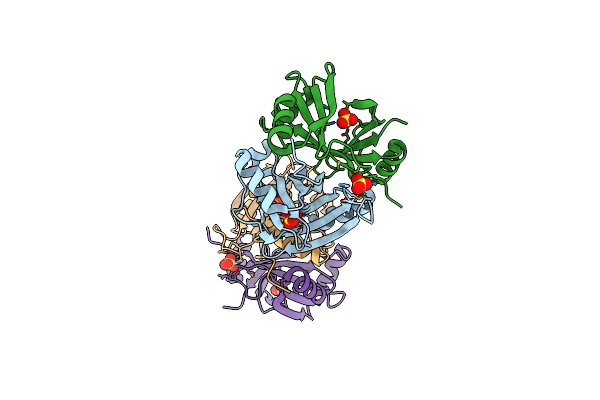
Crystal Structure Of Pyrococcus Furiosus 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2005-10-18 Classification: LYASE Ligands: CL, PEP, MN |
 |
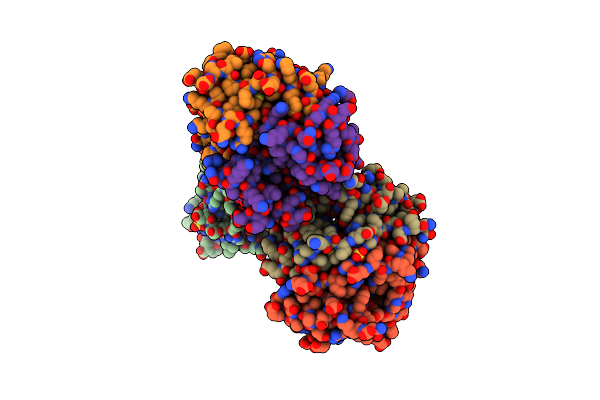
Organism: Propionibacterium freudenreichii subsp. shermanii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-07-11 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Propionibacterium freudenreichii subsp. shermanii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2001-07-11 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Bacillus caldovelox
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1999-04-16 Classification: HYDROLASE Ligands: MN |
 |
Organism: Bacillus caldovelox
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 1999-04-16 Classification: HYDROLASE Ligands: MN, GAI |
 |
Organism: Bacillus caldovelox
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1999-04-16 Classification: HYDROLASE Ligands: MN, ARG |
 |
Organism: Bacillus caldovelox
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 1999-04-16 Classification: HYDROLASE Ligands: MN, ORN, GAI |
 |
Organism: Bacillus caldovelox
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1999-04-16 Classification: HYDROLASE Ligands: MN, LYS, GAI |

