Search Count: 868
All
Selected
 |
Structure Of A Complex Between Pasteurella Multocida Surface Lipoprotein, Pmslp-1, And Bovine Complement Factor I
Organism: Pasteurella multocida 36950, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: NAG |
 |
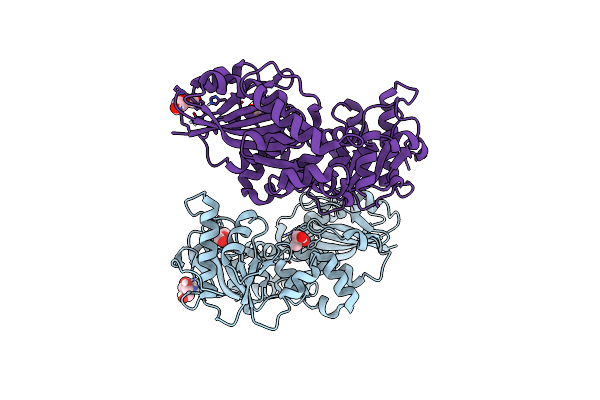
Crystal Structure Of A Slam-Dependent Surface Lipoprotein, Pmslp, In Pasteurella Multocida
Organism: Pasteurella multocida 36950
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: SE |
 |
Organism: Pasteurella multocida
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-09-25 Classification: SUGAR BINDING PROTEIN Ligands: TRS, PEG, GOL |
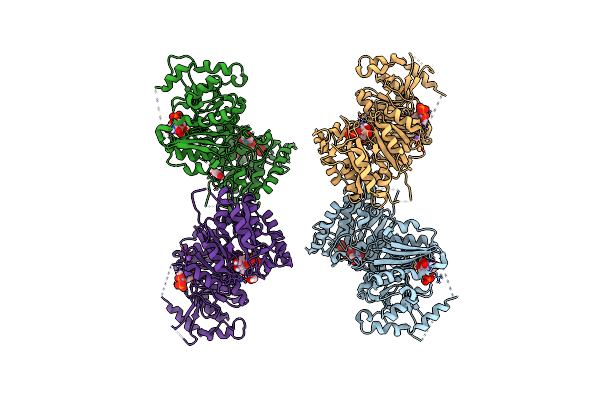 |
Crystal Structure Of Heparosan Synthase 2 From Pasteurella Multocida At 2.85 A
Organism: Pasteurella multocida
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2024-09-04 Classification: TRANSFERASE Ligands: MN, UDP, NA, CL, EDO, CA |
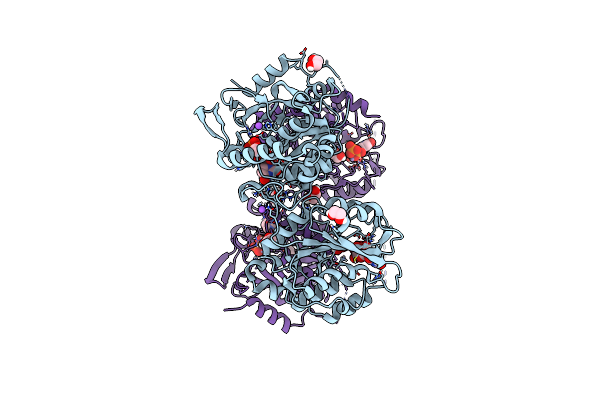 |
Crystal Structure Of Heparosan Synthase 2 From Pasteurella Multocida At 1.98 A
Organism: Pasteurella multocida
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: MN, UDP, EDO, NA |
 |
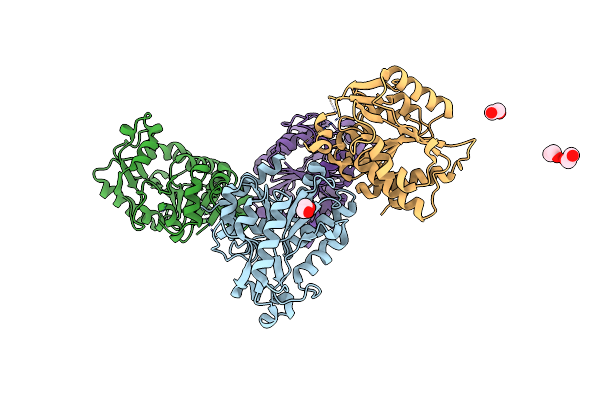
Cryo-Em Structure Of Heparosan Synthase 2 From Pasteurella Multocida With Polysaccharide In The Glcnac-T Active Site
Organism: Pasteurella multocida
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: MN, UDP |
 |
Organism: Pasteurella multocida
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2024-05-29 Classification: SUGAR BINDING PROTEIN Ligands: C5P, TRS, PGE |
 |
Organism: Pasteurella multocida (strain pm70)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-01-19 Classification: HYDROLASE Ligands: EDO |
 |
Organism: Pasteurella multocida (strain pm70)
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2020-09-09 Classification: HYDROLASE |
 |
Crystal Structure Of N-Acetylglucosamine-6-Phosphate Deacetylase From Pasteurella Multocida
Organism: Pasteurella multocida
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-03-04 Classification: METAL BINDING PROTEIN Ligands: ZN, PO4, EDO, GOL, NA |
 |
Crystal Structure Of N-Acetyl Mannosmaine Kinase In Complex With N-Acetylmannosamine In Pasteurella Multocida
Organism: Pasteurella multocida
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-02-05 Classification: SUGAR BINDING PROTEIN Ligands: BM3, ZN, GOL |
 |
Crystal Structure Of N-Acetyl Mannosmaine Kinase From Pasteurella Multocida
Organism: Pasteurella multocida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-02-05 Classification: SUGAR BINDING PROTEIN Ligands: ZN, GOL |
 |
Crystal Structure Of N-Acetyl Mannosmaine Kinase With Amp-Pnp From Pasteurella Multocida
Organism: Pasteurella multocida
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-02-05 Classification: SUGAR BINDING PROTEIN Ligands: ANP, CA |
 |
Organism: Pasteurella multocida
Method: X-RAY DIFFRACTION Resolution:3.33 Å Release Date: 2018-08-01 Classification: TRANSPORT PROTEIN Ligands: ASC, CA |
 |
Solution Nmr Structure Of The Membrane Localization Domain From Pasteurella Multocida Toxin
|
 |
Organism: Pasteurella multocida subsp. gallicida p1059
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2014-07-09 Classification: SUGAR BINDING PROTEIN Ligands: SLB |
 |
Crystal Structure Of Pasteurella Multocida N-Acetyl-D-Neuraminic Acid Lyase
Organism: Pasteurella multocida subsp. gallicida
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-11-06 Classification: LYASE Ligands: PO4, EDO |
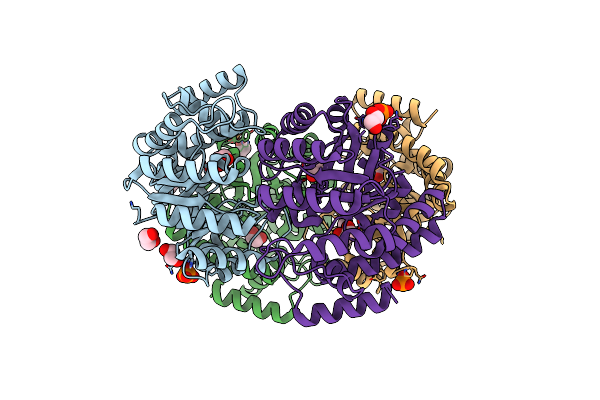 |
Crystal Structure Of Pasteurella Multocida N-Acetyl-D-Neuraminic Acid Lyase Trapped With Pyruvate Covalently Bound Through A Schiff Base To Lys164
Organism: Pasteurella multocida subsp. gallicida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-11-06 Classification: LYASE Ligands: PO4, EDO |
 |
Crystal Structure Of Pasteurella Multocida N-Acetyl-D-Neuraminic Acid Lyase K164A Mutant
Organism: Pasteurella multocida subsp. gallicida
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-11-06 Classification: LYASE Ligands: ME2, PG6 |
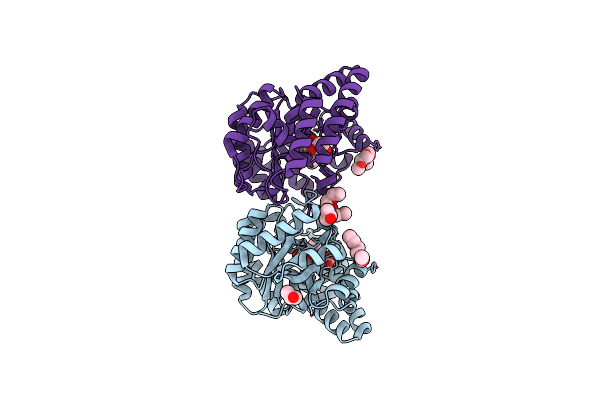 |
Crystal Structure Of Pasteurella Multocida N-Acetyl-D-Neuraminic Acid Lyase K164 Mutant Complexed With N-Acetylneuraminic Acid
Organism: Pasteurella multocida subsp. gallicida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-11-06 Classification: LYASE Ligands: SI3, ME2, CL, IPA |

